4RY0
 
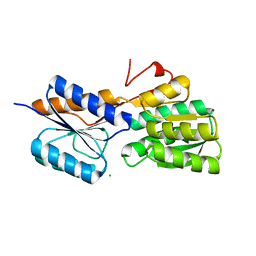 | | Crystal structure of ribose transporter solute binding protein RHE_PF00037 from Rhizobium etli CFN 42, TARGET EFI-511357, in complex with D-ribose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Probable ribose ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribose Transporter Solute Binding Protein from Rhizobium Etly, Target Efi-511357
To be Published
|
|
2OZ3
 
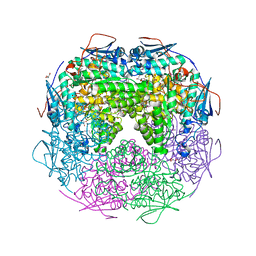 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
4RK7
 
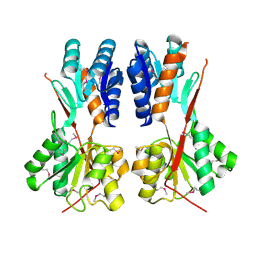 | | Crystal structure of LacI family transcriptional regulator CCPA from Weissella paramesenteroides, Target EFI-512926, with bound sucrose | | Descriptor: | Glucose-resistance amylase regulator, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of transcriptional regulator CCPA from Weissella paramesenteroides, Target EFI-512926
To be Published
|
|
4RXT
 
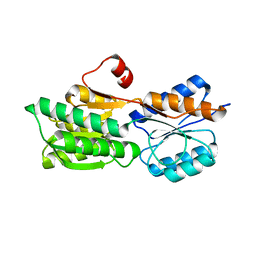 | | Crystal structure of carbohydrate transporter solute binding protein Arad_9553 from Agrobacterium Radiobacter, Target EFI-511541, in complex with D-arabinose | | Descriptor: | Sugar ABC transporter, alpha-L-arabinopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Sugar Binding Transporter Arad_9553 from Agrobacterium Radiobacter, Target Efi-511541
To be Published
|
|
3NF2
 
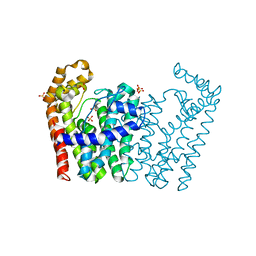 | | Crystal structure of polyprenyl synthetase from Streptomyces coelicolor A3(2) | | Descriptor: | Putative polyprenyl synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GRN
 
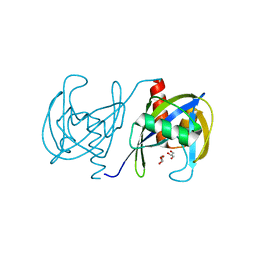 | | CRYSTAL STRUCTURE OF MUTT PROTEIN FROM Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, MutT related protein | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Do, J, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF MUTT PROTEIN FROM Methanosarcina mazei Go1
To be Published
|
|
3QAN
 
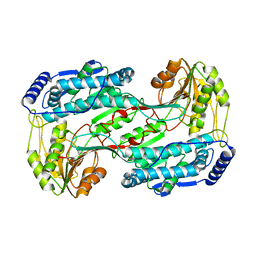 | | Crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from bacillus halodurans | | Descriptor: | 1-pyrroline-5-carboxylate dehydrogenase 1, ACETATE ION | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-01-11 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 1-Pyrroline-5-Carboxylate Dehydrogenase from Bacillus Halodurans
To be Published
|
|
2QDD
 
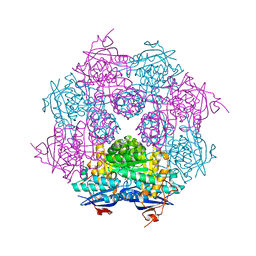 | | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Groshong, C, Gheyi, T, Sojitra, S, Wasserman, S.R, Koss, J, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM.
To be Published
|
|
3P9X
 
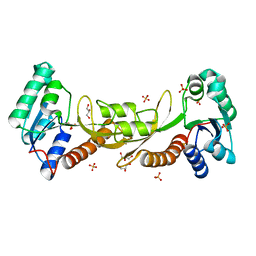 | | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyltransferase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-18 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans
To be Published
|
|
3OYR
 
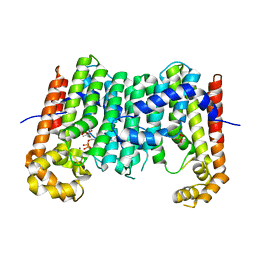 | | Crystal structure of polyprenyl synthase from Caulobacter crescentus CB15 complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3P8L
 
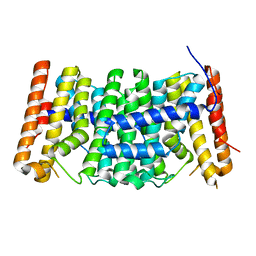 | | Crystal structure of polyprenyl synthase from Enterococcus faecalis V583 | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-14 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MOG
 
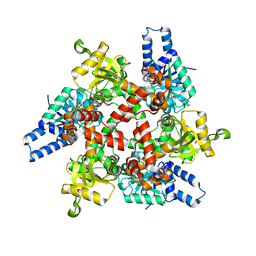 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3MZN
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | Descriptor: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N28
 
 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3N05
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
3N3D
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
2OOG
 
 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | Authors: | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
2OX4
 
 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
3PKO
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 complexed with citrate | | Descriptor: | CITRIC ACID, GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | Descriptor: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
3NFU
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | Descriptor: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3QU9
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with magnesium and tartrate | | Descriptor: | CHLORIDE ION, GLYCEROL, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3IK4
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RR1
 
 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J | | Descriptor: | CHLORIDE ION, D-MALATE, Putative D-galactonate dehydratase | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
3IRV
 
 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
