6J23
 
 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
1I34
 
 | | SOLUTION DNA QUADRUPLEX WITH DOUBLE CHAIN REVERSAL LOOP AND TWO DIAGONAL LOOPS CONNECTING GGGG TETRADS FLANKED BY G-(T-T) TRIAD AND T-T-T TRIPLE | | Descriptor: | 5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*CP*AP*GP*GP*GP*TP*TP*TP*TP*GP*GP*T)-3' | | Authors: | Kuryavyi, V, Majumdar, A, Shallop, A, Chernichenko, N, Skripkin, E, Jones, R, Patel, D.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple.
J.Mol.Biol., 310, 2001
|
|
1PIK
 
 | | ESPERAMICIN A1-DNA COMPLEX, NMR, 4 STRUCTURES | | Descriptor: | 2,4-dideoxy-3-O-methyl-4-(propan-2-ylamino)-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-S-methyl-4-thio-beta-D-ribo-hexopyranose, 2-deoxy-alpha-L-fucopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Esperamicin A1-DNA Complex
J.Mol.Biol., 265, 1997
|
|
1JO1
 
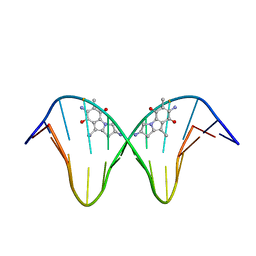 | | N7-Guanine Adduct of 2,7-diaminomitosene with DNA | | Descriptor: | 5'-D(*GP*TP*GP*(DAJ)GP*TP*AP*TP*AP*CP*CP*AP*C)-3', DECARBAMOYL-2,7-DIAMINOMITOSENE | | Authors: | Subramaniam, G, Paz, M.M, Kumar, G.S, Das, A, Palom, Y, Clement, C.C, Patel, D.J, Tomasz, M. | | Deposit date: | 2001-07-26 | | Release date: | 2001-09-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity.
Biochemistry, 40, 2001
|
|
1L4S
 
 | |
6IV5
 
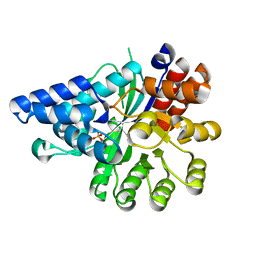 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
1JJP
 
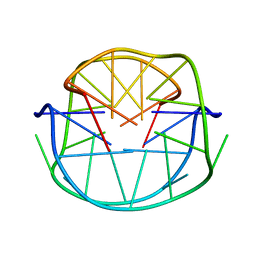 | | A(GGGG) Pentad-Containing Dimeric DNA Quadruplex Involving Stacked G(anti)G(anti)G(anti)G(syn) Tetrads | | Descriptor: | 5'-D(*GP*GP*GP*AP*GP*GP*TP*TP*TP*GP*GP*GP*AP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads.
J.Mol.Biol., 311, 2001
|
|
1JVC
 
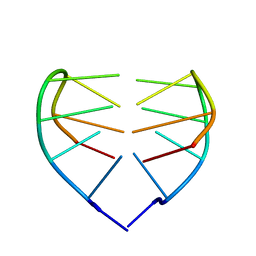 | | Dimeric DNA Quadruplex Containing Major Groove-Aligned A.T.A.T and G.C.G.C Tetrads Stabilized by Inter-Subunit Watson-Crick A:T and G:C Pairs | | Descriptor: | 5'-D(*GP*AP*GP*CP*AP*GP*GP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-08-29 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs.
J.Mol.Biol., 312, 2001
|
|
4NH3
 
 | |
4NT2
 
 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
2CWO
 
 | |
4ONJ
 
 | | Crystal structure of the catalytic domain of ntDRM | | Descriptor: | DNA methyltransferase, SINEFUNGIN | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases.
Cell(Cambridge,Mass.), 157, 2014
|
|
4ONQ
 
 | |
4NTO
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NT1
 
 | | Crystal structure of apo-form of Arabidopsis ACD11 (accelerated-cell-death 11) at 1.8 Angstrom resolution | | Descriptor: | SODIUM ION, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NH5
 
 | |
4NGC
 
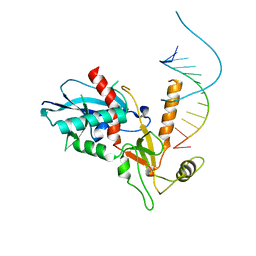 | |
4NGG
 
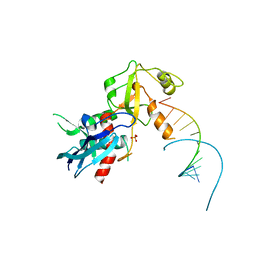 | |
4NH6
 
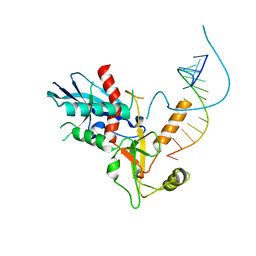 | |
4NGF
 
 | |
4QEP
 
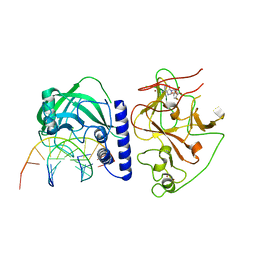 | | crystal structure of KRYPTONITE in complex with mCHG DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4QLM
 
 | | ydao riboswitch binding to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, RNA (108-MER), ... | | Authors: | Ren, A.M, Patel, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.721 Å) | | Cite: | c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets.
Nat.Chem.Biol., 10, 2014
|
|
4QBQ
 
 | | Crystal structure of DNMT3a ADD domain bound to H3 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone H3, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2014-05-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
To be Published
|
|
4QEN
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA and SAH | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*AP*TP*GP*AP*GP*TP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*TP*(5CM)P*AP*TP*CP*AP*GP*TP*AP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
