2JSQ
 
 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 15BrG, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
1JVC
 
 | | Dimeric DNA Quadruplex Containing Major Groove-Aligned A.T.A.T and G.C.G.C Tetrads Stabilized by Inter-Subunit Watson-Crick A:T and G:C Pairs | | Descriptor: | 5'-D(*GP*AP*GP*CP*AP*GP*GP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-08-29 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs.
J.Mol.Biol., 312, 2001
|
|
2JSL
 
 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 Natural, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-08 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
6O73
 
 | | Crystal structure of apo Csm1-Csm4 cassette | | Descriptor: | Csm1, Csm4, NICKEL (II) ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
5DDO
 
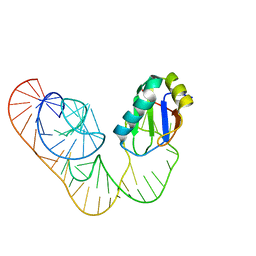 | |
6O79
 
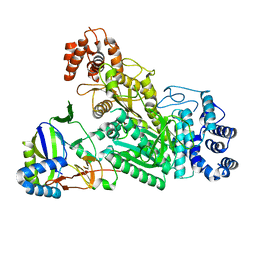 | | Crystal structure of Csm1-Csm4 cassette in complex with cA3 | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, cyclic RNA cA3 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
4EN5
 
 | | Crystal structure of fluoride riboswitch, Tl-Acetate soaked | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4ENA
 
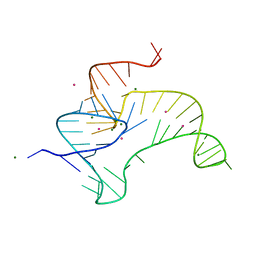 | | Crystal structure of fluoride riboswitch, soaked in Cs+ | | Descriptor: | CESIUM ION, FLUORIDE ION, Fluoride riboswitch, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
1XU1
 
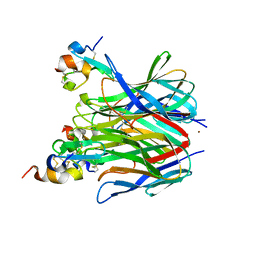 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
4ENB
 
 | | Crystal structure of fluoride riboswitch, bound to Iridium | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
1FJ5
 
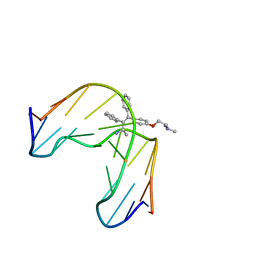 | | TAMOXIFEN-DNA ADDUCT | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINIUM, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Shimotakahara, S, Gorin, A, Kolbanovskiy, A, Kettani, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N, Patel, D.J. | | Deposit date: | 2000-08-07 | | Release date: | 2000-09-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Accomodation of S-cis-tamoxifen-N(2)-guanine adduct within a bent and widened DNA minor groove.
J.Mol.Biol., 302, 2000
|
|
1CR3
 
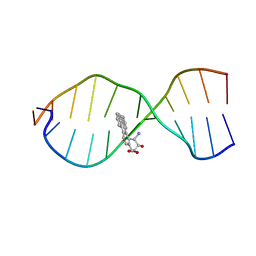 | | SOLUTION CONFORMATION OF THE (+)TRANS-ANTI-BENZO[G]CHRYSENE-DA ADDUCT OPPOSITE DT IN A DNA DUPLEX | | Descriptor: | BENZO[G]CHRYSENE, DNA (5'-D(*CP*TP*CP*TP*CP*AP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*TP*GP*AP*GP*AP*G)-3') | | Authors: | Suri, A.K, Mao, B, Amin, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1999-08-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex.
J.Mol.Biol., 292, 1999
|
|
2L81
 
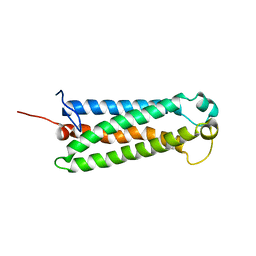 | | Solution NMR Structure of the serine-rich domain of hEF1 (Enhancer of filamentation 1) from Homo sapiens, Northeast Structural Genomics Consortium Target HR5554A | | Descriptor: | Enhancer of filamentation 1 | | Authors: | Liu, G, Xiao, R, Huang, Y.J, Patel, D, Ciccosanti, C.T, Acton, T.B, Tong, S, Everett, J.T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR5554A
To be Published
|
|
1XU2
 
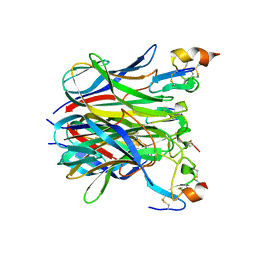 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
2KU7
 
 | |
3LMA
 
 | | Crystal structure of the stage V sporulation protein AD (SpoVAD) from Bacillus licheniformis. Northeast Structural Genomics Consortium Target BiR6. | | Descriptor: | Stage V sporulation protein AD (SpoVAD) | | Authors: | Vorobiev, S, Ashok, S, Seetharaman, J, Belote, R.L, Ciccosanti, C, Patel, D.J, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Crystal structure of the stage V sporulation protein AD (SpoVAD) from Bacillus licheniformis.
To be Published
|
|
4QXC
 
 | |
6VBW
 
 | |
6V9P
 
 | | Crystal structure of the transposition protein TniQ | | Descriptor: | GLYCEROL, TniQ family protein, ZINC ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
4QXB
 
 | |
4QXR
 
 | | Crystal structure of hSTING(S162A/G230I/Q266I) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
4QX8
 
 | |
5KK5
 
 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | Authors: | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | Deposit date: | 2016-06-21 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.289 Å) | | Cite: | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
4QXQ
 
 | | Crystal structure of hSTING(S162A/Q266I) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
4QXH
 
 | |
