1IW1
 
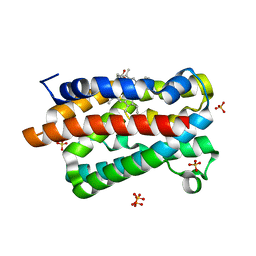 | | Crystal structure of a heme oxygenase (HmuO) from Corynebacterium diphtheriae complexed with heme in the ferrous state | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Hirotsu, S, Unno, M, Chu, G.C, Lee, D.S, Park, S.Y, Shiro, Y, Ikeda-Saito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-04 | | Release date: | 2003-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae.
J.Biol.Chem., 279, 2004
|
|
1UEK
 
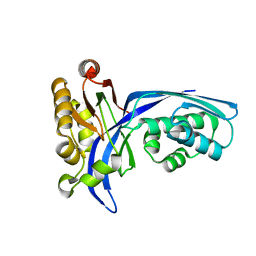 | | Crystal structure of 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Descriptor: | 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Authors: | Wada, T, Kuramitsu, S, Yokoyama, S, Tame, J.R.H, Park, S.Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis.
J.Biol.Chem., 278, 2003
|
|
6K6I
 
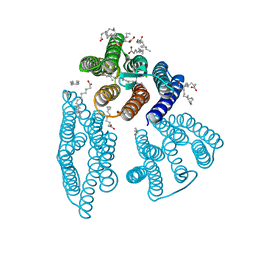 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
To Be Published
|
|
1V4W
 
 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH7.5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1VAT
 
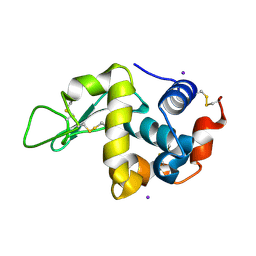 | | Iodine derivative of hen egg-white lysozyme | | Descriptor: | IODIDE ION, Lysozyme C | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
6K6K
 
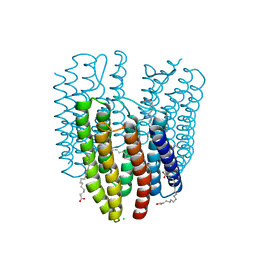 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
1V4X
 
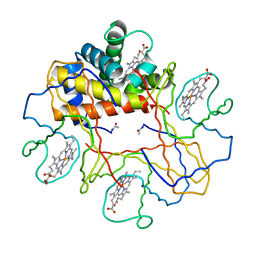 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH5.0 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V4U
 
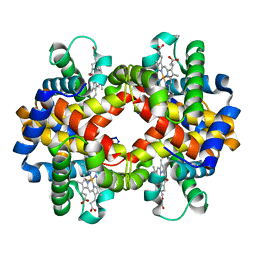 | | Crystal structure of bluefin tuna carbonmonoxy-hemoglobin | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, ... | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
6K6J
 
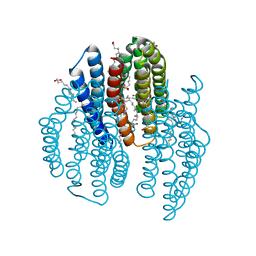 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | Descriptor: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
1VAU
 
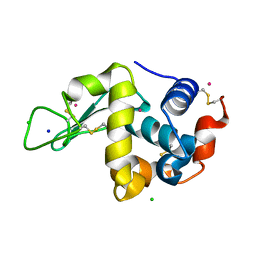 | | Xenon derivative of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
2IHS
 
 | |
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYB
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
5ZTK
 
 | | Synchrotron structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
6JY9
 
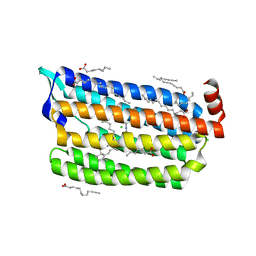 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY8
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY7
 
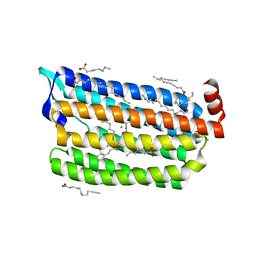 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
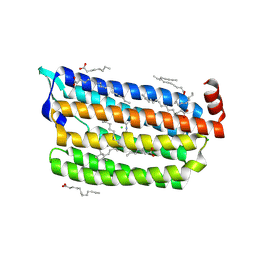 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
7VDN
 
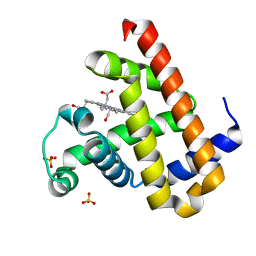 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
4NEE
 
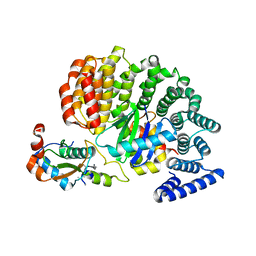 | | crystal structure of AP-2 alpha/simga2 complex bound to HIV-1 Nef | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit sigma, Protein Nef | | Authors: | Hurley, J.H, Bonifacino, J.S, Ren, X, Park, S.Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8841 Å) | | Cite: | How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4.
Elife, 3, 2014
|
|
7WJ5
 
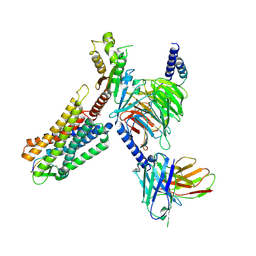 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
