1LLT
 
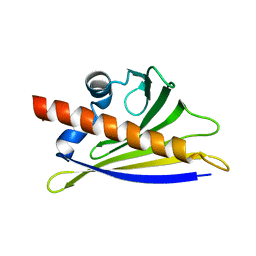 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT E45S | | Descriptor: | POLLEN ALLERGEN BET V 1 | | Authors: | Spangfort, M.D, Mirza, O, Ipsen, H, Van Neerven, R.J, Gajhede, M, Larsen, J.N. | | Deposit date: | 2002-04-30 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis.
J.Immunol., 171, 2003
|
|
6EYO
 
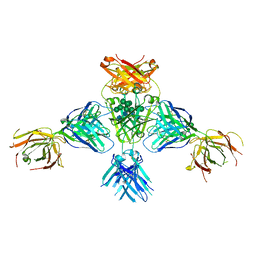 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
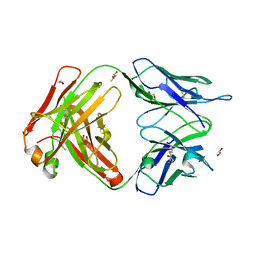 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
2KM2
 
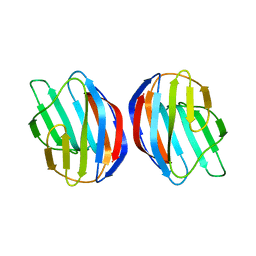 | | Galectin-1 dimer | | Descriptor: | Galectin-1 | | Authors: | Nesmelova, I.V, Ermakova, E, Daragan, V.A, Pang, M, Baum, L.G, Mayo, K.H. | | Deposit date: | 2009-07-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity.
J.Mol.Biol., 397, 2010
|
|
4J6K
 
 | |
4J6Q
 
 | |
5LGK
 
 | | Crystal structure of the human IgE-Fc bound to its B cell receptor derCD23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Dhaliwal, B, Pang, M.O.Y, Sutton, B.J. | | Deposit date: | 2016-07-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | IgE binds asymmetrically to its B cell receptor CD23.
Sci Rep, 7, 2017
|
|
4J6N
 
 | |
4KI1
 
 | | Primitive triclinic crystal form of the human IgE-Fc(epsilon)3-4 bound to its B cell receptor derCD23 | | Descriptor: | IG EPSILON CHAIN C REGION, LOW AFFINITY IMMUNOGLOBULIN EPSILON FC RECEPTOR, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Dhaliwal, B, Pang, M.O.Y, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2013-05-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A range of C3-C4 interdomain angles in IgE Fc accommodate binding to its receptor CD23.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5G64
 
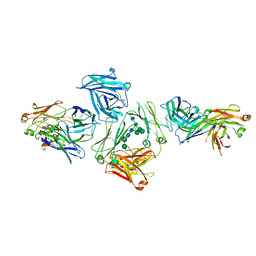 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | Descriptor: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.715 Å) | | Cite: | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
5LGJ
 
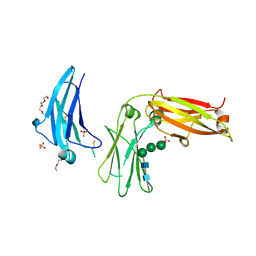 | |
4A5W
 
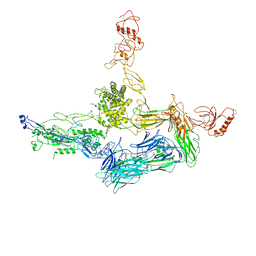 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
1FSK
 
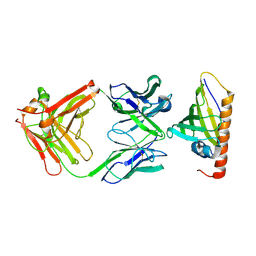 | | COMPLEX FORMATION BETWEEN A FAB FRAGMENT OF A MONOCLONAL IGG ANTIBODY AND THE MAJOR ALLERGEN FROM BIRCH POLLEN BET V 1 | | Descriptor: | ANTIBODY HEAVY CHAIN FAB, IMMUNOGLOBULIN KAPPA LIGHT CHAIN, MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Mirza, O, Henriksen, A, Ipsen, H, Larsen, J, Wissenbach, M, Spangfort, M, Gajhede, M. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-02 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1.
J.Immunol., 165, 2000
|
|
2JGX
 
 | | Structure of CCP module 7 of complement factor H - The AMD Not at risk varient (402Y) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
2JGW
 
 | | Structure of CCP module 7 of complement factor H - The AMD at risk varient (402H) | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
1QMR
 
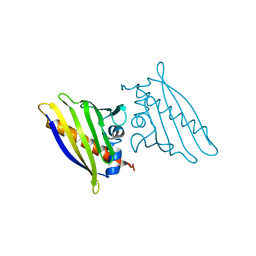 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | Descriptor: | MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
1XWV
 
 | | Structure of the house dust mite allergen Der f 2: Implications for function and molecular basis of IgE cross-reactivity | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Der f II | | Authors: | Johannessen, B.R, Skov, L.K, Kastrup, J.S, Kristensen, O, Bolwig, C, Larsen, J.N, Spangfort, M, Lund, K, Gajhede, M. | | Deposit date: | 2004-11-02 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the house dust mite allergen Der f 2: implications for function and molecular basis of IgE cross-reactivity.
Febs Lett., 579, 2005
|
|
3SNL
 
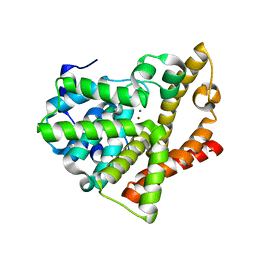 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SNI
 
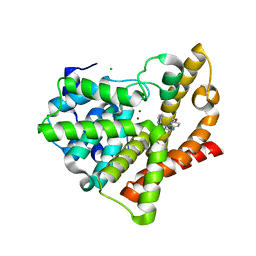 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
1BTV
 
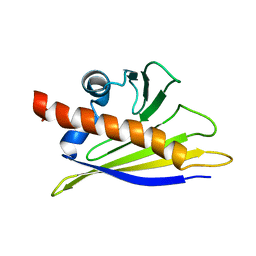 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1BV1
 
 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | BET V 1 | | Authors: | Gajhede, M, Osmark, P, Poulsen, F.M, Ipsen, H, Larson, J.N, Joostvan, R.J, Schou, C, Lowenstein, H, Spangfort, M.D. | | Deposit date: | 1997-07-08 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
2ATM
 
 | | Crystal structure of the recombinant allergen Ves v 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hyaluronoglucosaminidase, SULFATE ION | | Authors: | Skov, L.K, Seppala, U, Coen, J.J.F, Crickmore, N, King, T.P, Monsalve, R, Kastrup, J.S, Spangfort, M.D, Gajhede, M. | | Deposit date: | 2005-08-25 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant Ves v 2 at 2.0 Angstrom resolution: structural analysis of an allergenic hyaluronidase from wasp venom.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BZM
 
 | | Solution structure of the primary host recognition region of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Herbert, A.P, Uhrin, D, Lyon, M, Pangburn, M.K, Barlow, P.N. | | Deposit date: | 2005-08-18 | | Release date: | 2006-03-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Disease-Associated Sequence Variations Congregate in a Polyanion Recognition Patch on Human Factor H Revealed in Three-Dimensional Structure.
J.Biol.Chem., 281, 2006
|
|
2RLQ
 
 | | NMR structure of CCP modules 2-3 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-29 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
2RLP
 
 | | NMR structure of CCP modules 1-2 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-28 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
