5LIJ
 
 | | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Descriptor: | polyalanine chain built in bacteriophage phi812K1-420 cement protein density map | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6TBA
 
 | | Virion of native gene transfer agent (GTA) particle | | Descriptor: | IRON/SULFUR CLUSTER, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6I45
 
 | | Crystal structure of I13V/I62V/V77I South African HIV-1 subtype C protease containing a D25A mutation | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease, SODIUM ION | | Authors: | Sherry, D, Pandian, R, Achilonu, I.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-active site mutations in the HIV protease: Diminished drug binding affinity is achieved through modulating the hydrophobic sliding mechanism.
Int.J.Biol.Macromol., 217, 2022
|
|
6TO8
 
 | | Neck of empty GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Portal protein Rcc01684 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TEA
 
 | | Tail of native GTA particle computed with helical refinement, C6 symmetry | | Descriptor: | Phage major tail protein, TP901-1 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TEH
 
 | | Baseplate of native GTA particle computed with C3 symmetry | | Descriptor: | IRON/SULFUR CLUSTER, Putative gene transfer agent protein | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TE8
 
 | | Neck of native GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage portal protein, HK97 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TEB
 
 | | Tail-baseplate interface of native GTA particle computed with C6 symmetry | | Descriptor: | Distal tail protein Rcc01695, Tail tube protein Rcc01691 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TOA
 
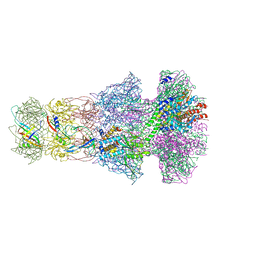 | | Neck of empty GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Portal protein Rcc01684, Stopper protein Rcc01689, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TE9
 
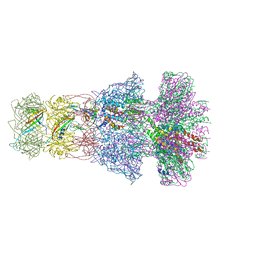 | | Neck of native GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage major tail protein, TP901-1 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TSV
 
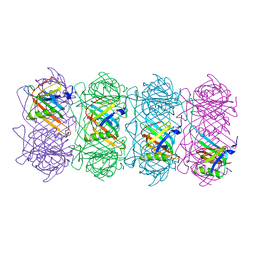 | | Tail of empty GTA particle computed with helical refinement, C6 symmetry | | Descriptor: | Tail tube protein Rcc01691 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TSW
 
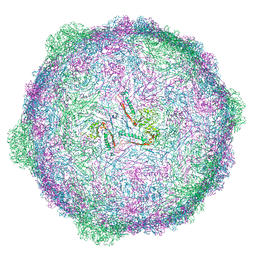 | | Isometric capsid of empty GTA particle computed with I4(I,n25r) symmetry | | Descriptor: | Major capsid protein Rcc01687 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
1YMP
 
 | |
1YR3
 
 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | SULFATE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-03 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene
J.Mol.Biol., 348, 2005
|
|
1YQU
 
 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | GUANINE, PHOSPHATE ION, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene
J.Mol.Biol., 348, 2005
|
|
2Q10
 
 | | RESTRICTION ENDONUCLEASE BcnI (WILD TYPE)-COGNATE DNA SUBSTRATE COMPLEX | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3'), ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2007-05-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Monomeric Restriction Endonuclease BcnI in the Apo Form and in an Asymmetric Complex with Target DNA.
J.Mol.Biol., 369, 2007
|
|
3WT4
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
1YQQ
 
 | | Escherichia coli purine nucleoside phosphorylase II, the product of the xapA gene | | Descriptor: | GUANINE, PHOSPHATE ION, Xanthosine phosphorylase | | Authors: | Dandanell, G, Szczepanowski, R.H, Kierdaszuk, B, Shugar, D, Bochtler, M. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli Purine Nucleoside Phosphorylase II, the Product of the xapA Gene
J.Mol.Biol., 348, 2005
|
|
5NYW
 
 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
1MSC
 
 | |
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
2V31
 
 | | Structure of First Catalytic Cysteine Half-domain of mouse ubiquitin- activating enzyme | | Descriptor: | UBIQUITIN-ACTIVATING ENZYME E1 X | | Authors: | Jaremko, L, Jaremko, M, Wojciechowski, W, Filipek, R, Szczepanowski, R.H, Bochtler, M, Zhukov, I. | | Deposit date: | 2007-06-11 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of First Catalytic Cysteine Half-Domain of Mouse Ubiquitin-Activating Enzyme
To be Published
|
|
1LMA
 
 | |
3IMB
 
 | | Alternative binding mode of restriction endonuclease BcnI to cognate DNA | | Descriptor: | 5'-D(*CP*GP*CP*CP*CP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*GP*GP*GP*CP*G)-3', R.BcnI | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Siksnys, V, Bochtler, M. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | How BcnI and MvaI distinguish W from S
To be Published
|
|
