3PKJ
 
 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3CKL
 
 | | Crystal structure of human cytosolic sulfotransferase SULT1B1 in complex with PAP and resveratrol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, RESVERATROL, Sulfotransferase family cytosolic 1B member 1 | | Authors: | Pan, P.W, Tempel, W, Dong, A, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-16 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human cytosolic sulfotransferase SULT1B1 in complex with PAP and resveratrol.
To be Published
|
|
3F3Y
 
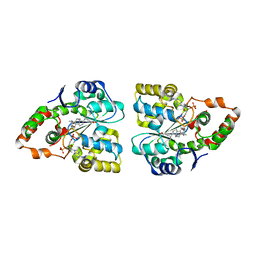 | | Crystal structure of human cytosolic sulfotransferase SULT2A1 in complex with PAP and lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Bile salt sulfotransferase | | Authors: | Pan, P.W, Dong, A, Amaya, M, Edwards, A.M. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | rystal structure of human cytosolic sulfotransferase SULT2A1 in complex with PAP and lithocholic acid
To be Published
|
|
4S3I
 
 | |
6RWO
 
 | | SIVrcm intasome (Q148H/G140S) in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6HN9
 
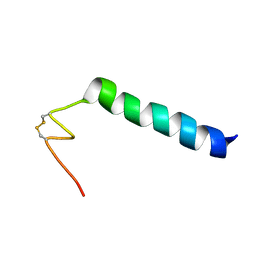 | | Nicomicin-1 -- Novel antimicrobial peptides from the Arctic polychaeta Nicomache minor provide new molecular insight into biological role of the BRICHOS domain | | Descriptor: | Nicomicin-1 | | Authors: | Panteleev, P.V, Tsarev, A.V, Bolosov, I.A, Paramonov, A.S, Marggraf, M.B, Sychev, S.V, Shenkarev, Z.O, Ovchinnikova, T.V. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Antimicrobial Peptides from the Arctic PolychaetaNicomache minorProvide New Molecular Insight into Biological Role of the BRICHOS Domain.
Mar Drugs, 16, 2018
|
|
5G4Q
 
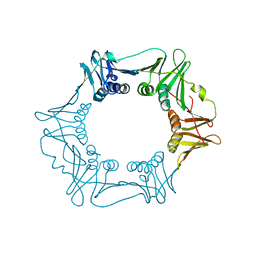 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
5G48
 
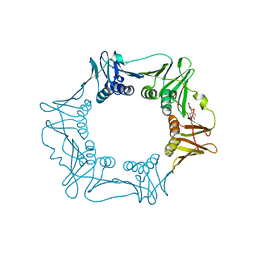 | | H.pylori Beta clamp in complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
5FVE
 
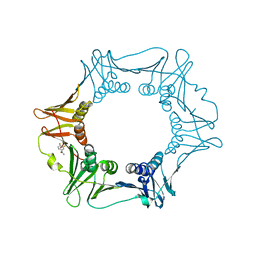 | |
5MMP
 
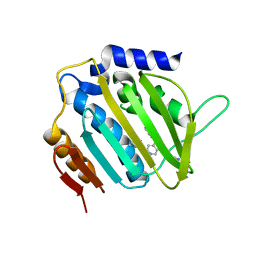 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMN
 
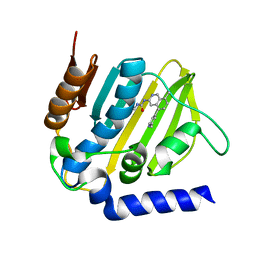 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMO
 
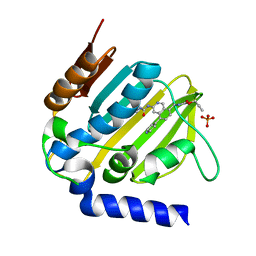 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with [3-(3-ethyl-ureido)-5-(pyridin-4-yl)-isoquinolin-8-yl-methyl]-carbamic acid prop-2-ynyl ester | | Descriptor: | DNA gyrase subunit B, PHOSPHATE ION, prop-2-ynyl ~{N}-[[3-(ethylcarbamoylamino)-5-pyridin-4-yl-isoquinolin-8-yl]methyl]carbamate | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5FXT
 
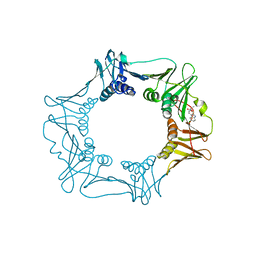 | |
6GX9
 
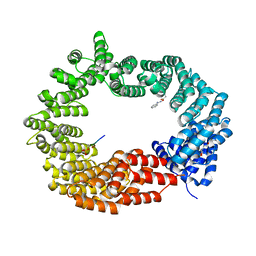 | | Crystal structure of the TNPO3 - CPSF6 RSLD complex | | Descriptor: | BENZAMIDINE, BICINE, Cleavage and polyadenylation specificity factor subunit 6, ... | | Authors: | Cherepanov, P, Cook, N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Nucleic Acids Res., 47, 2019
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z9E
 
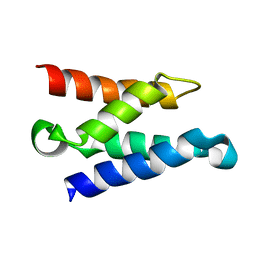 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | Descriptor: | PC4 and SFRS1 interacting protein 2 | | Authors: | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | Deposit date: | 2005-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
1GMY
 
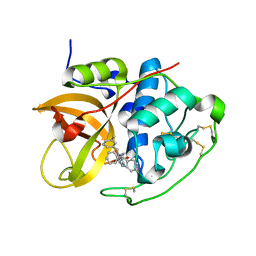 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
5FRQ
 
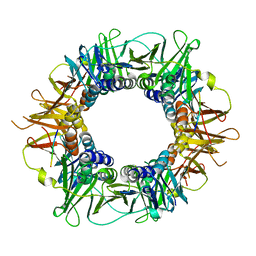 | |
4PO5
 
 | | Crystal structure of allophycocyanin B from Synechocystis PCC 6803 | | Descriptor: | Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, PHYCOCYANOBILIN, ... | | Authors: | Pang, P.P, Dong, L.L, Sun, Y.F, Zeng, X.L, Ding, W.L, Scheer, H, Yang, X, Zhao, K.H. | | Deposit date: | 2014-02-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DNE
 
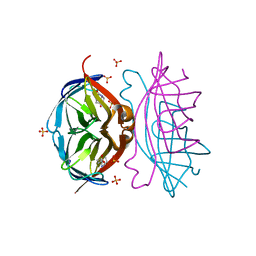 | | Crystal structure of a triple-mutant of streptavidin in complex with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, SULFATE ION, Streptavidin | | Authors: | Panwar, P, Deniaud, A, Pebay-Peyroula, E. | | Deposit date: | 2012-02-08 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Contamination from an affinity column: an encounter with a new villain in the world of membrane-protein crystallization.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
