8HFN
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7b | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(6-methoxypyridin-3-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFM
 
 | | Crystal Structure of Mycobacterium smegmatis MshC | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, ZINC ION | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFO
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7d | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(thiophen-2-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
6H9X
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with the Intermediate Analog 5'-O-(N-(L-Seryl)-Sulfamoyl)Adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acylated sulfonamide adenosines as potent inhibitors of the adenylate-forming enzyme superfamily.
Eur.J.Med.Chem., 174, 2019
|
|
8JC5
 
 | |
8JCA
 
 | |
5WQ4
 
 | | Crystal structure of OPTN and linear diubiquitin complex | | Descriptor: | Optineurin, ubiquitin | | Authors: | Li, F, Pan, L. | | Deposit date: | 2016-11-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation.
Autophagy, 14, 2018
|
|
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X81
 
 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X80
 
 | |
7X7Z
 
 | |
7XFR
 
 | |
4XKL
 
 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
2KBQ
 
 | |
3PVL
 
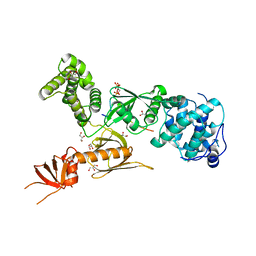 | | Structure of myosin VIIa MyTH4-FERM-SH3 in complex with the CEN1 of Sans | | Descriptor: | GLYCEROL, Myosin VIIa isoform 1, PHOSPHATE ION, ... | | Authors: | Wu, L, Pan, L.F, Wei, Z.Y, Zhang, M.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo.
Science, 331, 2011
|
|
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
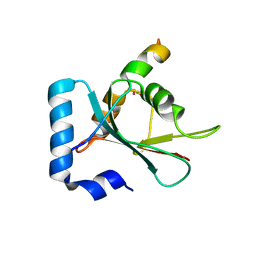 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3PZD
 
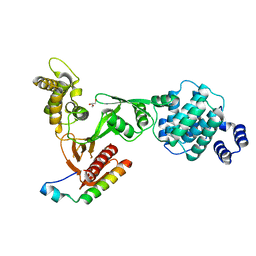 | | Structure of the myosin X MyTH4-FERM/DCC complex | | Descriptor: | GLYCEROL, Myosin-X, Netrin receptor DCC | | Authors: | Wei, Z, Yan, J, Pan, L, Zhang, M. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SHU
 
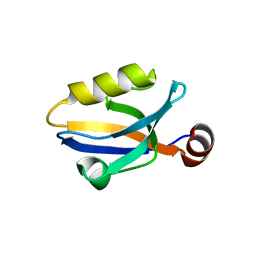 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
7YJ0
 
 | |
8W6A
 
 | |
8W6B
 
 | |
8X8K
 
 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
