4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4PM5
 
 | | Crystal structure of CTX-M-14 S70G beta-lactamase in complex with cefotaxime at 1.26 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
8SJ3
 
 | |
4PM8
 
 | | Crystal structure of CTX-M-14 S70G:S237A beta-lactamase at 1.17 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.169 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PMA
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase at 1.39 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Cardenas, A.M, Adamski, C.J, Brown, N.G, Horton, L.B, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM9
 
 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase in complex with cefotaxime at 1.45 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Cardenas, A.M, Adamski, C.J, Sankaran, B, Brown, N.G, Horton, L.B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM7
 
 | | Crystal structure of CTX-M-14 S70G:S237A in complex with cefotaxime at 1.29 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4PM6
 
 | | Crystal structure of CTX-M-14 S70G beta-lactamase at 1.56 Angstroms resolution | | Descriptor: | Beta-lactamase CTX-M-14, SULFATE ION | | Authors: | Adamski, C.J, Cardenas, A.M, Sankaran, B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
4RVA
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for deacylation | | Descriptor: | BICARBONATE ION, Beta-lactamase TEM | | Authors: | Stojanoski, V, Chow, D.-C, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4397 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX3
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, CITRATE ANION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-21 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7K5V
 
 | | OXA-48 bound by Compound 3.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-09-17 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7K2W
 
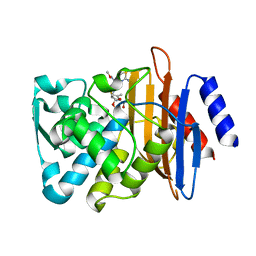 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7K2Y
 
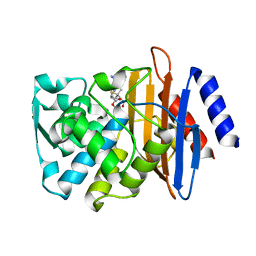 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7K2X
 
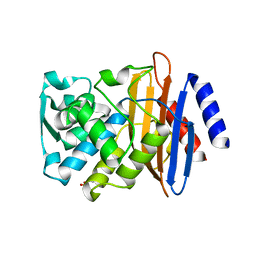 | | Crystal structure of CTX-M-14 E166A/K234R Beta-lactamase | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Lu, S, Palzkill, T, Sankaran, B, Hu, L, Soeung, V, Prasad, B.V.V. | | Deposit date: | 2020-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.
J.Biol.Chem., 295, 2020
|
|
7L8O
 
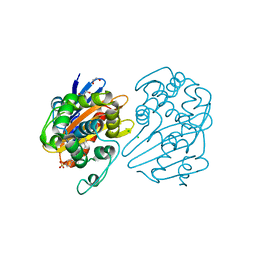 | | OXA-48 bound by Compound 4.3 | | Descriptor: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
