8SW5
 
 | |
4JS9
 
 | | Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Mesoheme, Nitric oxide synthase, ... | | Authors: | Hannibal, L, Page, R.C, Bolisetty, K, Yu, Z, Misra, S, Stuehr, D.J. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Kinetic and Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme
To be Published
|
|
3O9X
 
 | | Structure of the E. coli antitoxin MqsA (YgiT/b3021) in complex with its gene promoter | | Descriptor: | DNA (26-MER), GLYCEROL, Uncharacterized HTH-type transcriptional regulator ygiT, ... | | Authors: | Brown, B.L, Peti, W, Page, R. | | Deposit date: | 2010-08-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.0999 Å) | | Cite: | Structure of the Escherichia coli Antitoxin MqsA (YgiT/b3021) Bound to Its Gene Promoter Reveals Extensive Domain Rearrangements and the Specificity of Transcriptional Regulation.
J.Biol.Chem., 286, 2011
|
|
4KBO
 
 | | Crystal structure of the human Mortalin (GRP75) ATPase domain in the apo form | | Descriptor: | SODIUM ION, Stress-70 protein, mitochondrial | | Authors: | Amick, J, Page, R.C, Nix, J.C, Misra, S. | | Deposit date: | 2013-04-23 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone.
Protein Sci., 23, 2014
|
|
6VRO
 
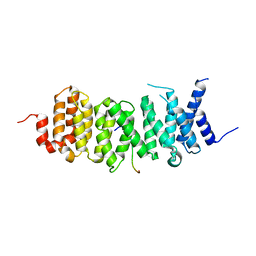 | | The structure of the PP2A B56 subunit AIM1 complex | | Descriptor: | Beta/gamma crystallin domain-containing protein 1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2020-02-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
6OBR
 
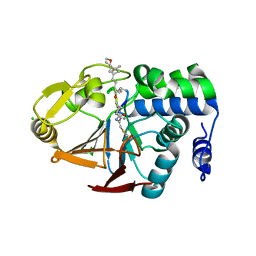 | | PP1 Y134A in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Microcystin LR, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBS
 
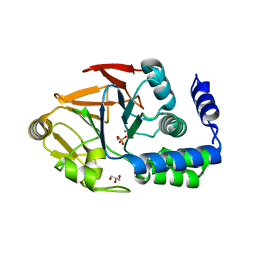 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5INB
 
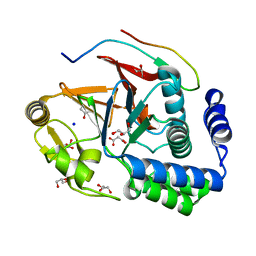 | | RepoMan-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5IOH
 
 | | RepoMan-PP1a (protein phosphatase 1, alpha isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
2QDC
 
 | |
2QDP
 
 | |
6P2U
 
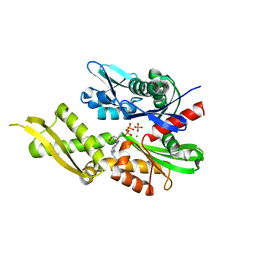 | | Structure of Mortalin-NBD with N6-propargyladenosine-5'-diphosphate | | Descriptor: | 9-{5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-alpha-D-ribofuranosyl}-N-(prop-2-yn-1-yl)-9H-purin-6-amine, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Moseng, M.A, Page, R.C. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2- and N6-functionalized adenosine-5'-diphosphate analogs for the inhibition of mortalin.
Febs Lett., 593, 2019
|
|
3EGH
 
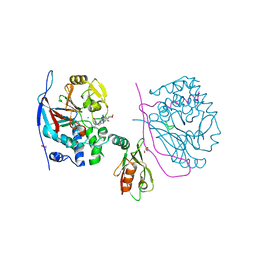 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6PMT
 
 | |
6OYL
 
 | | The structure of the PP2A B56 subunit KIF4A complex | | Descriptor: | Chromosome-associated kinesin KIF4A, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
6HKW
 
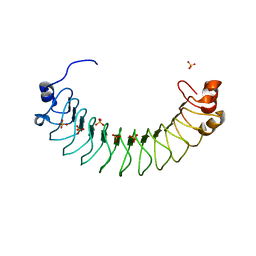 | | Crystal structure of human SDS22 | | Descriptor: | Protein phosphatase 1 regulatory subunit 7, SULFATE ION | | Authors: | Heroes, E, Choy, M.S, Page, R, Peti, W, Ulens, C, Van Meervelt, L, Nys, M, Bollen, M. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Guided Exploration of SDS22 Interactions with Protein Phosphatase PP1 and the Splicing Factor BCLAF1.
Structure, 27, 2019
|
|
3GA8
 
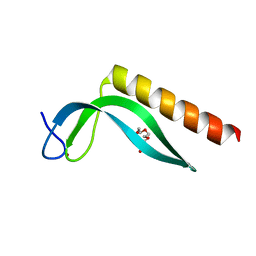 | | Structure of the N-terminal domain of the E. coli protein MqsA (YgiT/b3021) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, HTH-type transcriptional regulator MqsA (YgiT/B3021), ZINC ION | | Authors: | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
3HI2
 
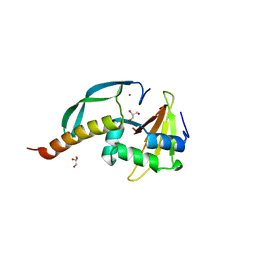 | | Structure of the N-terminal domain of the E. coli antitoxin MqsA (YgiT/b3021) in complex with the E. coli toxin MqsR (YgiU/b3022) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator mqsA(ygiT), Motility quorum-sensing regulator mqsR, ... | | Authors: | Grigoriu, S, Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
3E7B
 
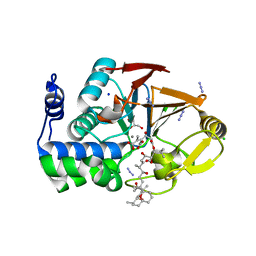 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10R)-10-{(2S,3S,6R,8S,9R)-3,9-dimethyl-8-[(3S)-3-methyl-4-oxopentyl]-1,7-dioxaspiro[5.5]undec-2-yl}-3,7-dihydroxy-2-methoxy-6-methyl-1-(1-methylethyl)-5-oxoundecyl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
3E7A
 
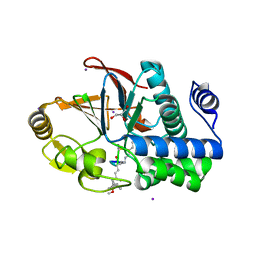 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin Nodularin-R | | Descriptor: | AZIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
3GN5
 
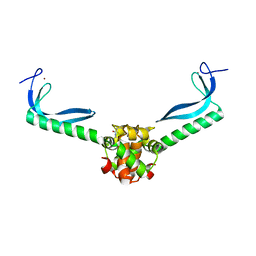 | | Structure of the E. coli protein MqsA (YgiT/b3021) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MQSA (YGIT/b3021), ZINC ION | | Authors: | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-03-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
7MOV
 
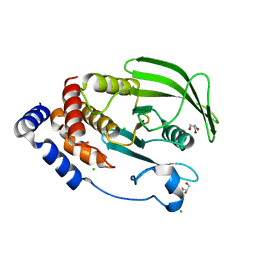 | | PTP1B 1-301 F225Y-R199N mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
4G9Y
 
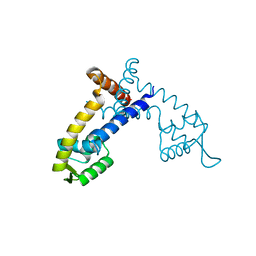 | | Crystal Structure of the PcaV transcriptional regulator from Streptomyces coelicolor | | Descriptor: | GLYCEROL, PcaV transcriptional regulator | | Authors: | Brown, B.L, Davis, J.R, Sello, J.K, Page, R. | | Deposit date: | 2012-07-24 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors.
Nucleic Acids Res., 41, 2013
|
|
4F0Z
 
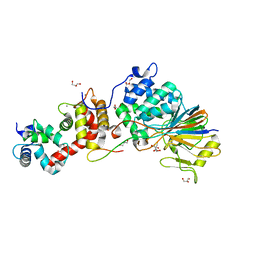 | | Crystal Structure of Calcineurin in Complex with the Calcineurin-Inhibiting Domain of the African Swine Fever Virus Protein A238L | | Descriptor: | Ankyrin repeat domain-containing protein A238L, CALCIUM ION, Calcineurin subunit B type 1, ... | | Authors: | Grigoriu, S, Peti, W, Page, R. | | Deposit date: | 2012-05-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin.
Plos Biol., 11, 2013
|
|
