2JNS
 
 | |
1WG0
 
 | | Structural comparison of Nas6p protein structures in two different crystal forms | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Nakamura, Y, Umehara, T, Tanaka, A, Horikoshi, M, Yokoyama, S, Padmanabhan, B, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural comparison of Nas6p protein structures in two different crystal forms
To be Published
|
|
5YTU
 
 | |
5YUL
 
 | | Native Structure of hSOD1 in P6322 space group | | Descriptor: | Dihydrogen tetrasulfide, GLYCEROL, Superoxide dismutase [Cu-Zn], ... | | Authors: | Manjula, R, Padmanabhan, B. | | Deposit date: | 2017-11-22 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assessment of ligand binding at a site relevant to SOD1 oxidation and aggregation
FEBS Lett., 592, 2018
|
|
6A9O
 
 | | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis | | Descriptor: | 2'-[(6-oxo-5,6-dihydrophenanthridin-3-yl)carbamoyl][1,1'-biphenyl]-2-carboxylic acid, DIMETHYL SULFOXIDE, Dihydrogen tetrasulfide, ... | | Authors: | Manjula, R, Padmanabhan, B. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis.
J.Biomol.Struct.Dyn., 37, 2019
|
|
1X0J
 
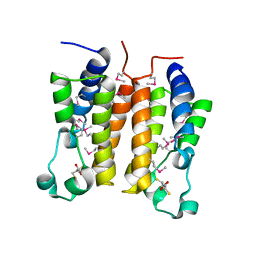 | | Crystal structure analysis of the N-terminal bromodomain of human Brd2 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-23 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for acetylated histone H4 recognition by the human Brd2 bromodomain
To be Published
|
|
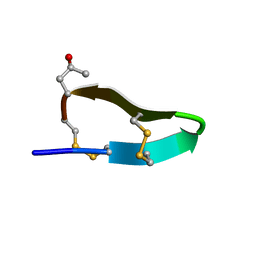
2M62
 
 | |
2DVW
 
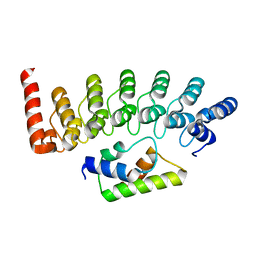 | |
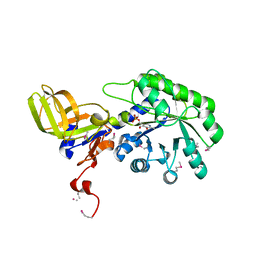
2YXX
 
 | |
2DVS
 
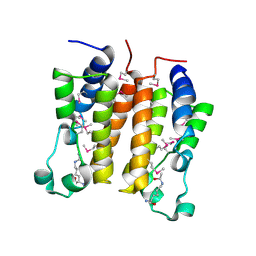 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVR
 
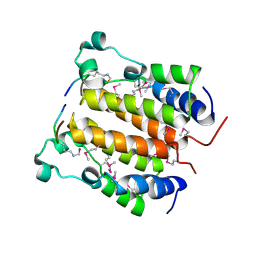 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
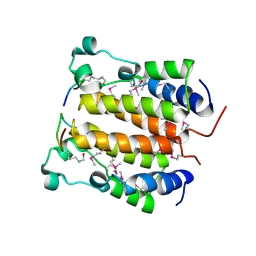 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DZO
 
 | |
2DZN
 
 | |
2YYZ
 
 | | Crystal structure of Sugar ABC transporter, ATP-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, Sugar ABC transporter, ... | | Authors: | Ethayathullah, A.S, Bessho, Y, Padmanabhan, B, Singh, T.P, Kaur, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of Sugar ABC transporter, ATP-binding protein
To be Published
|
|
2YZ2
 
 | | Crystal structure of the ABC transporter in the cobalt transport system | | Descriptor: | Putative ABC transporter ATP-binding protein TM_0222 | | Authors: | Ethayathullah, A.S, Bessho, Y, Padmanabhan, B, Singh, T.P, Kaur, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ABC transporter in the cobalt transport system
To be Published
|
|
2YXY
 
 | | Crystarl structure of Hypothetical conserved protein, GK0453 | | Descriptor: | Hypothetical conserved protein, GK0453 | | Authors: | Nakamu, Y, Bessho, Y, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the hypothetical DUF1811-family protein GK0453 from Geobacillus kaustophilus HTA426
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3AQA
 
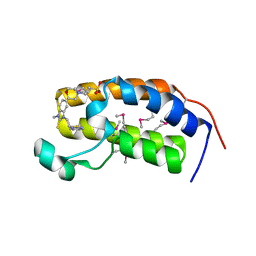 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
2YYY
 
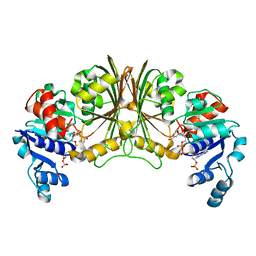 | | Crystal structure of Glyceraldehyde-3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malay, A.D, Bessho, Y, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from the archaeal hyperthermophile Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2YYR
 
 | |
2Z34
 
 | |
2Z3F
 
 | |
