2QSU
 
 | |
1TJW
 
 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant with bound argininosuccinate | | Descriptor: | ARGININOSUCCINATE, Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
2N8U
 
 | |
2NQG
 
 | | Calpain 1 proteolytic core inactivated by WR18(S,S), an epoxysuccinyl-type inhibitor. | | Descriptor: | 5-AZANYLIDYNE-N-[(2S)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]-L-NORVALYL-L-ARGINYL-L-TRYPTOPHANAMIDE, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
2MSI
 
 | |
2QTG
 
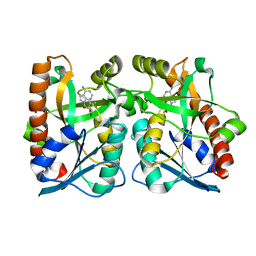 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with 5'-methylthiotubercidin | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine nucleosidase | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
2QTT
 
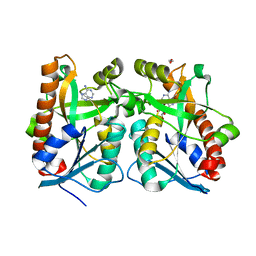 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 1,2-ETHANEDIOL, 5'-methylthioadenosine nucleosidase, ... | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
4MF3
 
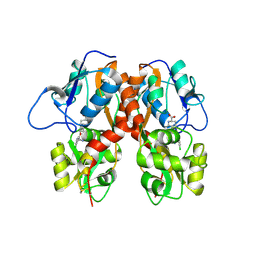 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4NK6
 
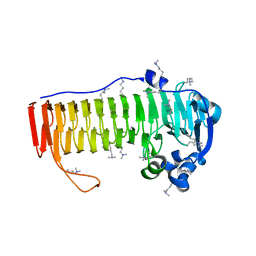 | |
4MSI
 
 | |
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4OZX
 
 | |
4OZZ
 
 | |
4OKH
 
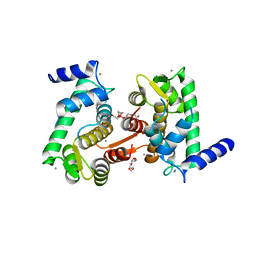 | | Crystal structure of calpain-3 penta-EF-hand domain | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CALCIUM ION, Calpain-3 | | Authors: | Karunan Partha, S, Ravulapalli, R, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of calpain-3 penta-EF-hand (PEF) domain - a homodimerized PEF family member with calcium bound at the fifth EF-hand.
Febs J., 281, 2014
|
|
4OCW
 
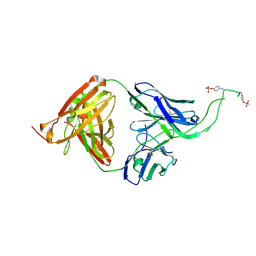 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4ODE
 
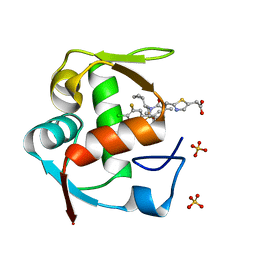 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | (2-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}-1,3-thiazol-5-yl)acetic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4ODH
 
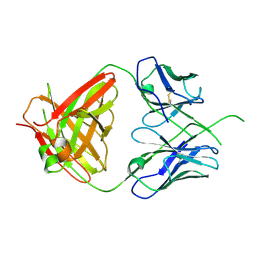 | | Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.UCA heavy chain, CAP256-VRC26.UCA light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OCS
 
 | | Crystal structure of human Fab CAP256-VRC26.10, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.10 heavy chain, CAP256-VRC26.10 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OZY
 
 | |
4OD3
 
 | | Crystal structure of human Fab CAP256-VRC26.07, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CAP256-VRC26.07 heavy chain, CAP256-VRC26.07 light chain, ... | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4M3O
 
 | | Crystal structure of K.lactis Rtr1 NTD | | Descriptor: | KLLA0F12672p, ZINC ION | | Authors: | Hsu, P.L, Yang, W, Zheng, N, Varani, G. | | Deposit date: | 2013-08-06 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rtr1 Is a Dual Specificity Phosphatase That Dephosphorylates Tyr1 and Ser5 on the RNA Polymerase II CTD.
J.Mol.Biol., 426, 2014
|
|
4ORG
 
 | | Crystal structure of human Fab CAP256-VRC26.04, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.04 heavy chain, CAP256-VRC26.04 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.121 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OZV
 
 | | Crystal Structure of the periplasmic alginate lyase AlgL | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid | | Authors: | Howell, P.L, Wolfram, F, Robinson, H, Arora, K. | | Deposit date: | 2014-02-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
J.Biol.Chem., 298, 2022
|
|
4OZW
 
 | |
