3M5G
 
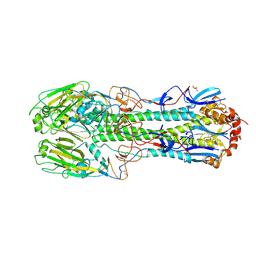 | | Crystal structure of a H7 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Chen, L.M, Carney, P.J, Donis, R.O, Stevens, J. | | Deposit date: | 2010-03-12 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site.
Plos Pathog., 6, 2010
|
|
3MP9
 
 | | Structure of Streptococcal protein G B1 domain at pH 3.0 | | Descriptor: | FORMIC ACID, Immunoglobulin G-binding protein G | | Authors: | Tomlinson, J.H, Green, V.L, Baker, P.J, Williamson, M.P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural origins of pH-dependent chemical shifts in the B1 domain of protein G.
Proteins, 78, 2010
|
|
4AGG
 
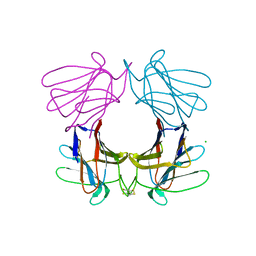 | | Structure of a tetrameric galectin from Cinachyrella sp. (Ball sponge) | | Descriptor: | CHLORIDE ION, GALECTIN | | Authors: | Freymann, D.M, Focia, P.J, Sakai, R, Swanson, G.T. | | Deposit date: | 2012-01-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of a Tetrameric Galectin from Cinachyrella Sp. (Ball Sponge).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3N0S
 
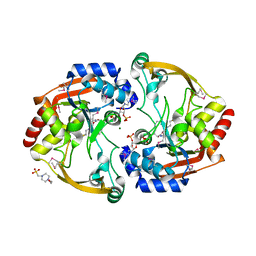 | | Crystal structure of BA2930 mutant (H183A) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
7JH3
 
 | | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE, DI(HYDROXYETHYL)ETHER | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP
To Be Published
|
|
4GKH
 
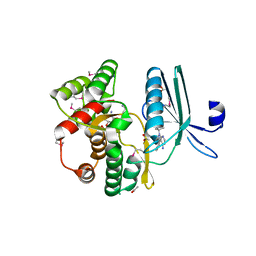 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
3M5H
 
 | | Crystal structure of a H7 influenza virus hemagglutinin complexed with 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yang, H, Chen, L.M, Carney, P.J, Donis, R.O, Stevens, J. | | Deposit date: | 2010-03-12 | | Release date: | 2010-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site.
Plos Pathog., 6, 2010
|
|
5UWK
 
 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound (S)-10a | | Descriptor: | (S)-3-methyl-2-(4'-(((4-oxo-4,5,6,7-tetrahydro-3H-cyclopenta[d]pyrimidin-2-yl)thio)methyl)-[1,1'-biphenyl]-4-ylsulfonamido)butanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Taylor, A.B, Cao, X, Hart, P.J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective Matrix Metalloproteinase 13 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
3N5N
 
 | |
4BKQ
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype)with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPZ3_3519 | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
4BL6
 
 | | Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to co-ordinate recruitment of cargos to dynein | | Descriptor: | ARGININE, PROTEIN BICAUDAL D | | Authors: | Liu, Y, Salter, H.K, Holding, A.N, Johnson, C.M, Stephens, E, Lukavsky, P.J, Walshaw, J, Bullock, S.L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-12 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bicaudal-D Uses a Parallel, Homodimeric Coiled Coil with Heterotypic Registry to Coordinate Recruitment of Cargos to Dynein
Genes Dev., 27, 2013
|
|
4FZS
 
 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
8BAD
 
 | | Tpp80Aa1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Binary toxin A-like protein, CALCIUM ION, ... | | Authors: | Best, H.L, Williamson, L.J, Rizkallah, P.J, Berry, C, Lloyd-Evans, E. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids.
Toxins, 14, 2022
|
|
4BGW
 
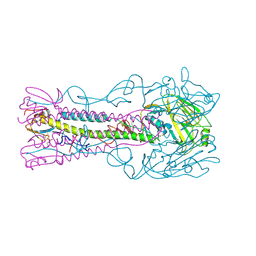 | | Crystal Structure of H5 (VN1194) Influenza Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
4BGY
 
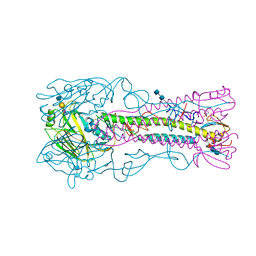 | | H5 (VN1194) Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
7K8N
 
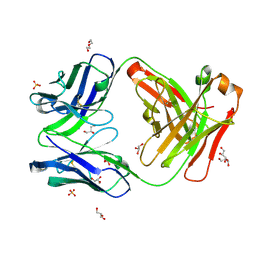 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8X
 
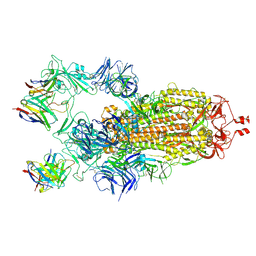 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Q
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
4BGX
 
 | | H5 (VN1194) Influenza Virus Haemagglutinin in Complex with Human Receptor Analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
7K8Y
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
4BKO
 
 | | Enoyl-ACP reducatase FabV from Burkholderia pseudomallei (apo) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PUTATIVE REDUCTASE BURPS305_1051, ... | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Point Mutation Changes Substrate Binding Mechanism and Inhibitor Specificity of Yersinia Pestis Enoyl- Acp Reductase Fabv
To be Published
|
|
3MKH
 
 | | Podospora anserina Nitroalkane Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NITROALKANE OXIDASE, ... | | Authors: | Tormos, J.R, Taylor, A.B, Daubner, S.C, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Identification of a hypothetical protein from Podospora anserina as a nitroalkane oxidase.
Biochemistry, 49, 2010
|
|
