1G6H
 
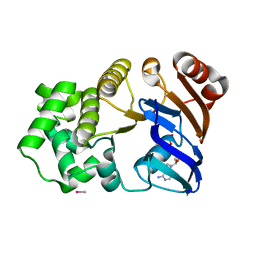 | | CRYSTAL STRUCTURE OF THE ADP CONFORMATION OF MJ1267, AN ATP-BINDING CASSETTE OF AN ABC TRANSPORTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HIGH-AFFINITY BRANCHED-CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Yuan, Y.-R, Martsinkevich, O, Karpowich, N, Millen, L, Thomas, P.J, Hunt, J.F. | | Deposit date: | 2000-11-06 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter.
Structure, 9, 2001
|
|
1GAJ
 
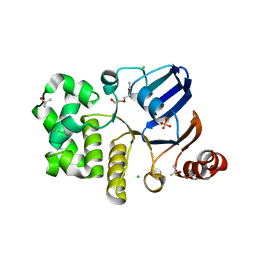 | | CRYSTAL STRUCTURE OF A NUCLEOTIDE-FREE ATP-BINDING CASSETTE FROM AN ABC TRANSPORTER | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HIGH-AFFINITY BRANCHED CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, ... | | Authors: | Karpowich, N, Yuan, Y.-R, Dai, P.L, Martsinkevich, O, Millen, L, Thomas, P.J, Hunt, J.F. | | Deposit date: | 2000-11-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter.
Structure, 9, 2001
|
|
1F85
 
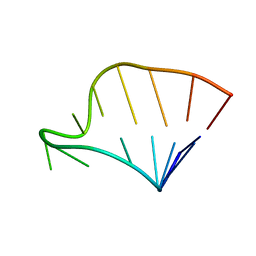 | | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIIE | | Descriptor: | HCV-1B IRES RNA DOMAIN IIIE | | Authors: | Lukavsky, P.J, Otto, G.A, Lancaster, A.M, Sarnow, P, Puglisi, J.D. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function.
Nat.Struct.Biol., 7, 2000
|
|
6U7Q
 
 | | NMR solution structure of SFTI-R10 | | Descriptor: | GLY-ARG-CYS-THR-LYS-SER-ILE-PRO-PRO-ARG-CYS-PHE-PRO-ASP inhibitor | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1FOU
 
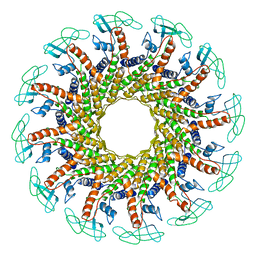 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S.N, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
8DFZ
 
 | |
6WAD
 
 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor | | Descriptor: | 5-bromo-N-(diphenylmethyl)-N-methylthiophene-2-carboxamide, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Schapira, M, De Freitas, R.F, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor
to be published
|
|
1FT0
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI K113A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6WAN
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6Q5T
 
 | |
6VU7
 
 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | Descriptor: | CHLORIDE ION, YbjN protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
1D9X
 
 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | Descriptor: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
6WAM
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6VL3
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VJH
 
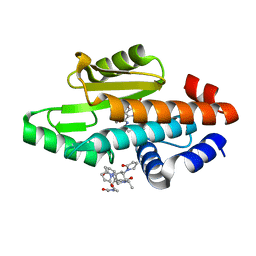 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VIV
 
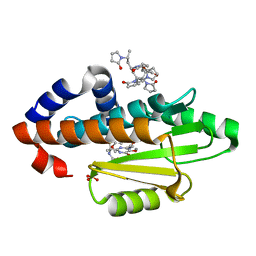 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3BRB
 
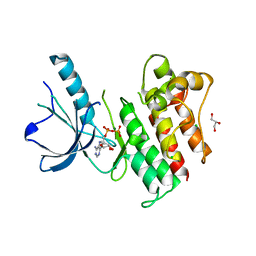 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
6W10
 
 | |
6VM6
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 2'3'3'-cAAA | | Descriptor: | 2'-5'-Linked Cyclic RNA (5'-R(P*AP*AP*A)-3'), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
1DQG
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE RICH DOMAIN OF MANNOSE RECEPTOR | | Descriptor: | MANNOSE RECEPTOR, SULFATE ION | | Authors: | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
3BBH
 
 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
7NZZ
 
 | |
3BBE
 
 | | M. jannaschii Nep1 | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
1GGZ
 
 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
