6ALL
 
 | | Crystal structure of a predicted ferric/iron (III) hydroxymate siderophore substrate binding protein from Bacillus anthracis | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Fe(3+)-citrate-binding protein yfmC | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of a predicted ferric/iron (III) hydroxymate siderophore substrate binding protein from Bacillus anthracis
To Be Published
|
|
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IHV
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IHW
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
2RKJ
 
 | | Cocrystal structure of a tyrosyl-tRNA synthetase splicing factor with a group I intron RNA | | Descriptor: | RNA (238-MER), RNA (5'-R(P*GP*CP*UP*U)-3'), Tyrosyl-tRNA synthetase | | Authors: | Paukstelis, P.J, Chen, J.-H, Chase, E, Lambowitz, A.M, Golden, B.L. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
Nature, 451, 2008
|
|
1JB2
 
 | | CRYSTAL STRUCTURE OF NTF2 M84E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JXJ
 
 | | Role of mobile loop in the mechanism of human salivary amylase | | Descriptor: | Alpha-amylase, salivary, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Wang, Z, Mishra, P.J, Thomas, L.M. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
7P5L
 
 | |
7P8G
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MALONATE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-07-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form
To Be Published
|
|
1JKU
 
 | | Crystal Structure of Manganese Catalase from Lactobacillus plantarum | | Descriptor: | CALCIUM ION, HYDROXIDE ION, MANGANESE (III) ION, ... | | Authors: | Barynin, V.V, Whittaker, M.M, Antonyuk, S.V, Lamzin, V.S, Harrison, P.M, Artymiuk, P.J, Whittaker, J.W. | | Deposit date: | 2001-07-13 | | Release date: | 2002-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of manganese catalase from Lactobacillus plantarum.
Structure, 9, 2001
|
|
7PD5
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid
To Be Published
|
|
1JJD
 
 | |
7PHX
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - acid-stable sulfotyrosine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J, Calisto, B.M. | | Deposit date: | 2021-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and evaluation of peptidic thrombin inhibitors bearing acid-stable sulfotyrosine analogues.
Chem.Commun.(Camb.), 57, 2021
|
|
1K42
 
 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
2VE1
 
 | | Isopenicillin N synthase with substrate analogue AsMCOV (oxygen exposed 1min 20bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
6AOJ
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal yeast Hog1p sequence | | Descriptor: | CHLORIDE ION, Ceg4, MAGNESIUM ION | | Authors: | Stogios, P.J, Nocek, B, Cuff, M.E, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
1HME
 
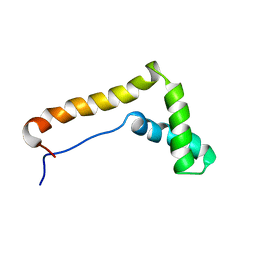 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
1HL5
 
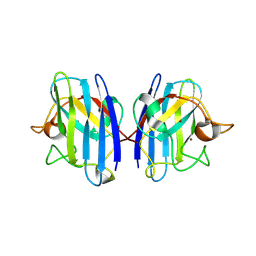 | | The Structure of Holo Type Human Cu, Zn Superoxide Dismutase | | Descriptor: | CALCIUM ION, COPPER (II) ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Holo and Metal-Deficient Wild-Type Human Cu, Zn Superoxide Dismutase and its Relevance to Familial Amyotrophic Lateral Sclerosis
J.Mol.Biol., 328, 2003
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
1JB4
 
 | | CRYSTAL STRUCTURE OF NTF2 M102E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JBO
 
 | | The 1.45A Three-Dimensional Structure of c-Phycocyanin from the Thermophylic Cyanobacterium Synechococcus elongatus | | Descriptor: | C-Phycocyanin alpha chain, C-Phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nield, J, Rizkallah, P.J, Barber, J, Chayen, N.E. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45A three-dimensional structure of C-phycocyanin from the
thermophilic cyanobacterium Synechococcus elongatus
J.STRUCT.BIOL., 141, 2003
|
|
7PPZ
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PQ0
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
