2FTC
 
 | | Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L12, ... | | Authors: | Mears, J.A, Sharma, M.R, Gutell, R.R, Richardson, P.E, Agrawal, R.K, Harvey, S.C. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | A Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome
J.Mol.Biol., 358, 2006
|
|
2FX9
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a thioether-linked peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein gp41 | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
3ICD
 
 | | STRUCTURE OF A BACTERIAL ENZYME REGULATED BY PHOSPHORYLATION, ISOCITRATE DEHYDROGENASE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Thorsness, P.E, Ramalingam, V, Helmers, N.H, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1989-12-28 | | Release date: | 1991-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2FX8
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41) | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
3INQ
 
 | | Crystal structure of BCL-XL in complex with W1191542 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(biphenyl-3-ylmethyl)piperazin-1-yl]-N-{[4-({(1R)-3-(dimethylamino)-1-[(phenylsulfanyl)methyl]propyl}amino)-3-nitrophenyl]sulfonyl}benzamide, Bcl-2-like protein 1, ... | | Authors: | Fairlie, W.D, Smith, B.J, Colman, P.M, Czabotar, P.E, Lee, E.F. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands
J. Biol. Chem., 284, 2009
|
|
6GNZ
 
 | |
6GO0
 
 | |
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
8TYF
 
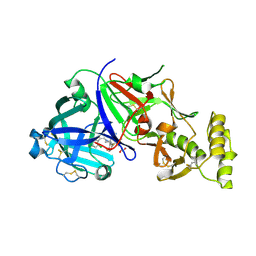 | | Plasmodium vivax PMV-WM06 inhibitor complex | | Descriptor: | (2E,4aR,7aS)-6-[(3M)-3-(2-chlorophenyl)pyridin-2-yl]-7a-(2,5-difluorophenyl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hodder, A.N, Scally, S.W, Czabotar, P.E, Cowman, A.F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Plasmodium vivax PMX-XX inhibitor complex
To Be Published
|
|
1XOA
 
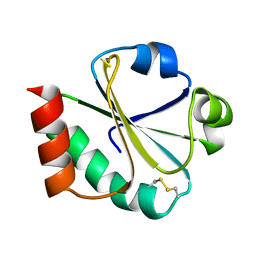 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
3IO8
 
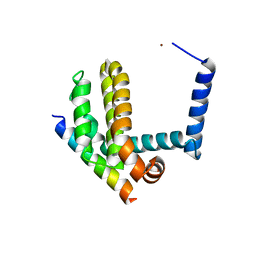 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
2HDP
 
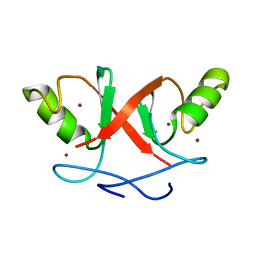 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
1ANT
 
 | |
1B3K
 
 | | Plasminogen activator inhibitor-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Sharp, A.M, Stein, P.E, Pannu, N.S, Read, R.J. | | Deposit date: | 1998-12-11 | | Release date: | 1999-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion.
Structure Fold.Des., 7, 1999
|
|
1XOB
 
 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1HZS
 
 | | Crystal structure of a peptide nucleic acid duplex (BT-PNA) containing a bicyclic analogue of thymine | | Descriptor: | PEPTIDE NUCLEIC ACID | | Authors: | Eldrup, A.B, Nielsen, B.B, Haaima, G, Rasmussen, H, Kastrup, J.S, Christensen, C, Nielsen, P.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,8-Naphthyridin-2(1H)-ones. Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs)
Eur.J.Org.Chem., 9, 2001
|
|
1AX3
 
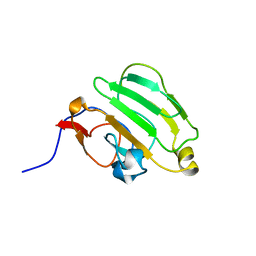 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1XJ9
 
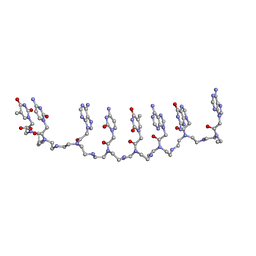 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | Descriptor: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | Authors: | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2004-09-23 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
1JVQ
 
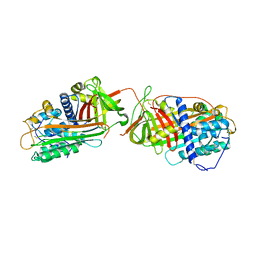 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1BN6
 
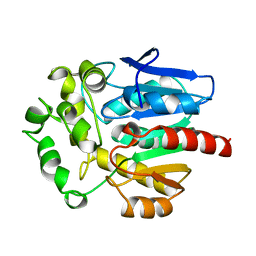 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1Y7J
 
 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
1Y7K
 
 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
1UUB
 
 | |
1BN7
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | ACETATE ION, HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
