8G1T
 
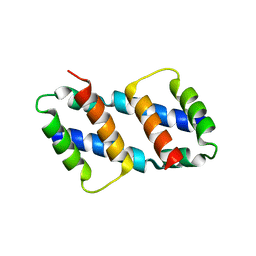 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
3NFB
 
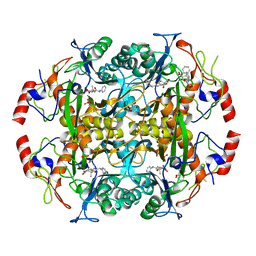 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with hydrolyzed ampicillin | | Descriptor: | (2S,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-peptidyl aminopeptidase, GLYCEROL, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
1COU
 
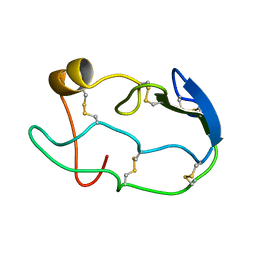 | |
8QKK
 
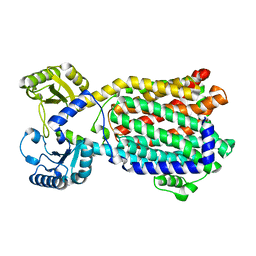 | | Cryo-EM structure of MmpL3 from Mycobacterium smegmatis reconstituted into peptidiscs | | Descriptor: | Trehalose monomycolate exporter MmpL3 | | Authors: | Couston, J, Guo, Z, Wang, K, Gourdon, P.E, Blaise, M. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the trehalose monomycolate transporter, MmpL3, reconstituted into peptidiscs.
Curr Res Struct Biol, 6, 2023
|
|
7SH1
 
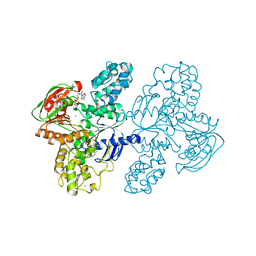 | | Class II UvrA protein - Ecm16 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Excinuclease ABC subunit UvrA, ... | | Authors: | Grade, P, Erlandson, A, Ullah, A, Mathews, I.I, Chen, X, Kim, C.-Y, Mera, P.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Sci Rep, 13, 2023
|
|
7OP3
 
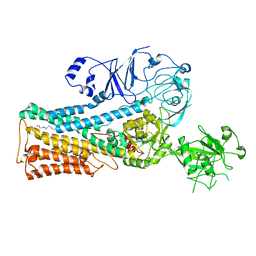 | | Cryo-EM structure of P5B-ATPase E2PiSPM | | Descriptor: | Cation-transporting ATPase, SPERMINE | | Authors: | Li, P, Gronberg, C, Wang, K.T, Salustros, N, Gourdon, P.E. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
2PCO
 
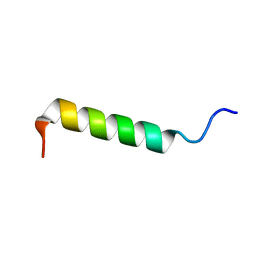 | | Spatial Structure and Membrane Permeabilization for Latarcin-1, a Spider Antimicrobial Peptide | | Descriptor: | Latarcin-1 | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity.
Biochemistry, 47, 2008
|
|
8GHR
 
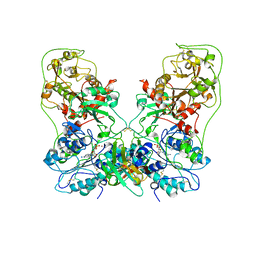 | | Structure of human ENPP1 in complex with variable heavy domain VH27.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Carozza, J.A, Wang, H, Solomon, P.E, Wells, J.A, Li, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of VH domains that allosterically inhibit ENPP1.
Nat.Chem.Biol., 20, 2024
|
|
4V2K
 
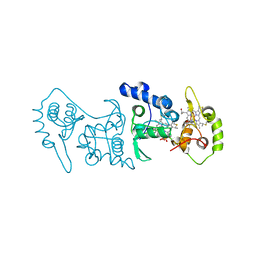 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | Descriptor: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | Authors: | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-10-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
4UWQ
 
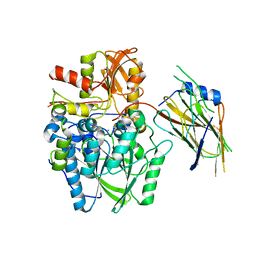 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | Descriptor: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | Authors: | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-08-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1KBH
 
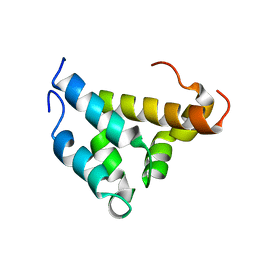 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
4URA
 
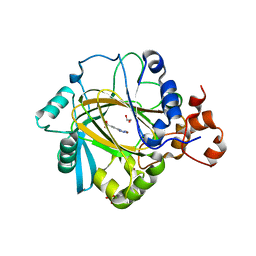 | | Crystal structure of human JMJD2A in complex with compound 14a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2H-1,2,3-triazol-4-yl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Krojer, T, England, K.S, Vollmar, M, Crawley, L, Williams, E, Riesebos, E, Szykowska, A, Burgess-Brown, N, Oppermann, U, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
Medchemcomm, 5, 2014
|
|
1U2N
 
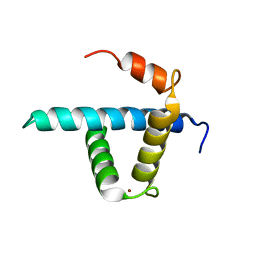 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
6WLH
 
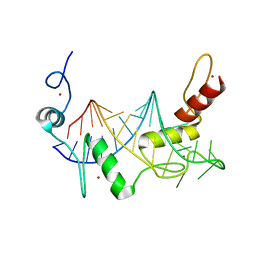 | |
4M6L
 
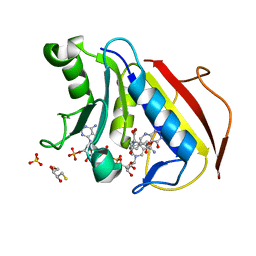 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M6J
 
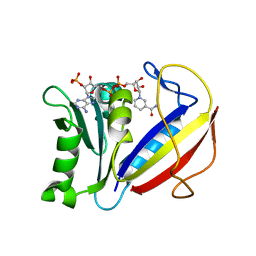 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADPH | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1TOT
 
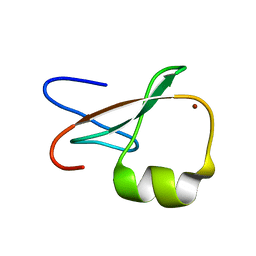 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
5JR2
 
 | | Crystal structure of the EphA4 LBD in complex with APYd3 peptide inhibitor | | Descriptor: | APYd3 peptide, Ephrin type-A receptor 4, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Olson, E.J, Pasquale, E.B, Dawson, P.E, Riedl, S.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modifications of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor To Achieve High Plasma Stability.
Acs Med.Chem.Lett., 7, 2016
|
|
1KDX
 
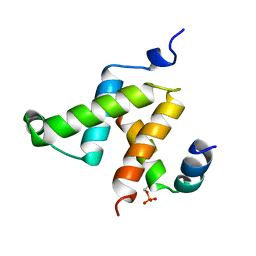 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
5L82
 
 | |
4XDL
 
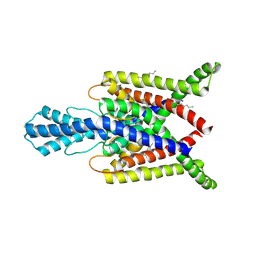 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with a brominated fluoxetine derivative. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[2-bromanyl-4-(trifluoromethyl)phenoxy]-N-methyl-3-phenyl-propan-1-amine, CADMIUM ION, ... | | Authors: | Mackenzie, A, Pike, A.C.W, Dong, Y.Y, Mukhopadhyay, S, Ruda, G.F, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
6X8O
 
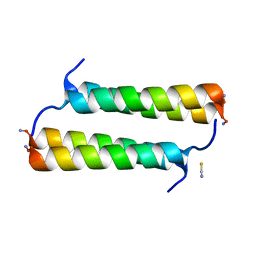 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
6O96
 
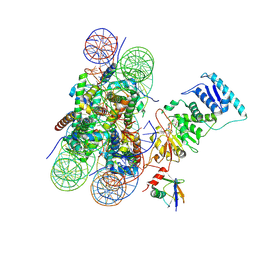 | | Dot1L bound to the H2BK120 Ubiquitinated nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Vasilyev, N, Chen, R, Nudler, E, Armache, J.-P, Armache, K.-J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination.
Mol.Cell, 74, 2019
|
|
6OM3
 
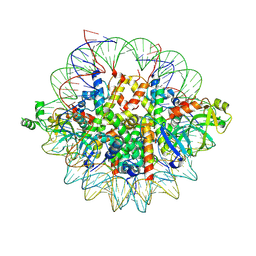 | | Crystal structure of the Orc1 BAH domain in complex with a nucleosome core particle | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A, ... | | Authors: | De Ioannes, P.E, Wang, M, Armache, K.-J. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of the Orc1 BAH-nucleosome complex.
Nat Commun, 10, 2019
|
|
4FAS
 
 | |
