8SS0
 
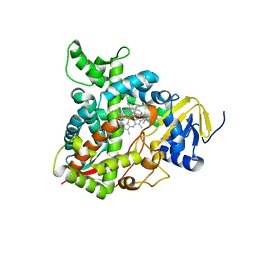 | | Human sterol 14 alpha-demethylase (CYP51) in complex with the reaction intermediate 14 alpha-aldehyde dihydrolanosterol | | Descriptor: | 3beta-hydroxy-10alpha,13alpha-lanosta-8,24-dien-30-al, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Guengerich, F.P, Lepesheva, G.I. | | Deposit date: | 2023-05-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-18 Labeling Reveals a Mixed Fe-O Mechanism in the Last Step of Cytochrome P450 51 Sterol 14 alpha-Demethylation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6UCE
 
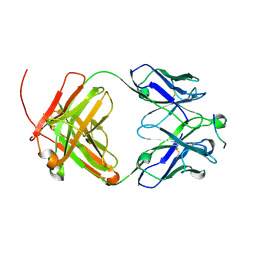 | |
4M7J
 
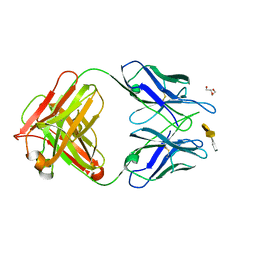 | | Crystal structure of S25-26 in complex with Kdo(2.8)Kdo(2.4)Kdo trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V, Muller-Loennies, S, Saldova, R, Muniyappa, M, Brade, L, Rudd, P.M, Harvey, D.J, Kosma, P, Brade, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Groove-type Recognition of Chlamydiaceae-specific Lipopolysaccharide Antigen by a Family of Antibodies Possessing an Unusual Variable Heavy Chain N-Linked Glycan.
J.Biol.Chem., 289, 2014
|
|
3NQ4
 
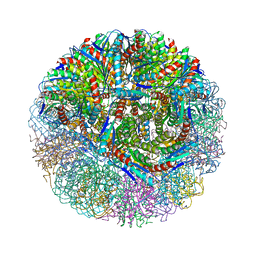 | |
6UO0
 
 | |
2DGC
 
 | |
6AIM
 
 | | Cab2 mutant H337A complex with phosphopantothenate-cysteine | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl-L-cysteine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
1R4L
 
 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
4MC3
 
 | | Hedycaryol synthase in complex with Nerolidol | | Descriptor: | (3R,6E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
6LU9
 
 | |
3NZM
 
 | |
4HIY
 
 | |
4Q3D
 
 | | PylD cocrystallized with L-Ornithine-Nd-D-ornithine and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7VT5
 
 | | Crystal structure of CBM deleted MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, METHIONINE, TRYPTOPHAN | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3RY4
 
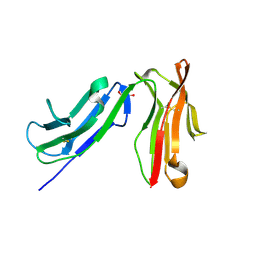 | |
7VT6
 
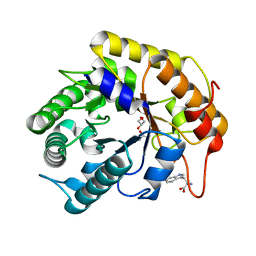 | | Crystal structure of CBM deleted MtGlu5 in complex with BGC. | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4HSO
 
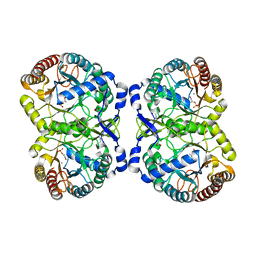 | | Crystal structure of S213G variant DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
6AI9
 
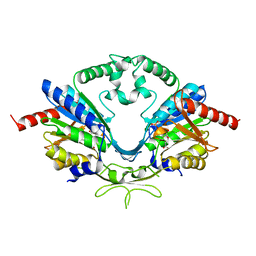 | | Cab2 mutant-H337A complex with phosphopantothenate | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
3EMP
 
 | |
6M7Y
 
 | |
4HOA
 
 | | Crystal structure of the complex of type 1 ribosome inactivating protein from Momordica Balsamina with B-D-galactopyranosyl-(1-4)-D-glucose at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of type 1 ribosome inactivating protein from Momordica Balsamina with B-D-galactopyranosyl-(1-4)-D-glucose at 2.0 A resolution
To be Published
|
|
6UCF
 
 | |
6M5T
 
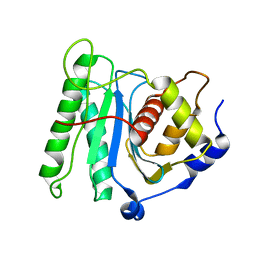 | | The coordinate of the nuclease domain of the apo terminase complex | | Descriptor: | Tripartite terminase subunit 3 | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
8CQ2
 
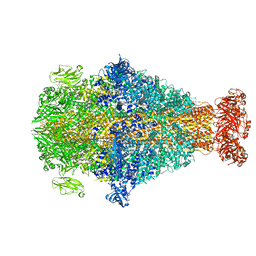 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
2DTU
 
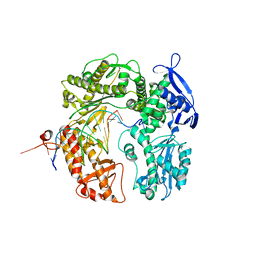 | | Crystal structure of the beta hairpin loop deletion variant of RB69 gp43 in complex with DNA containing an abasic site analog | | Descriptor: | 5'-D(*CP*GP*(3DR)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*GP*A)-3', DNA polymerase | | Authors: | Aller, P, Hogg, M, Konigsberg, W, Wallace, S.S, Doublie, S. | | Deposit date: | 2006-07-15 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family.
J.Biol.Chem., 282, 2007
|
|
