7T4B
 
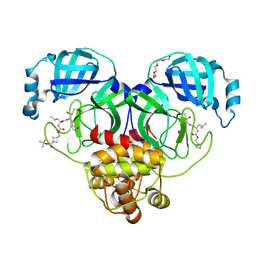 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T42
 
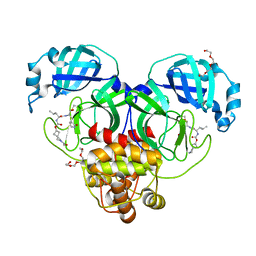 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
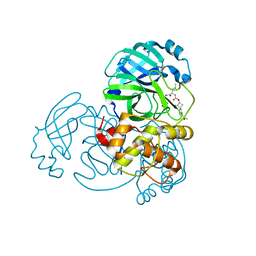 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T45
 
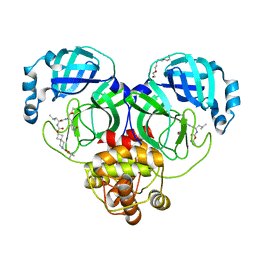 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 7c | | Descriptor: | (1S,2S)-2-{[N-({[7-(tert-butoxycarbonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Kankanamalage, A.C.G, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
1JP6
 
 | |
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T44
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
1Y9T
 
 | |
1JUJ
 
 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
7T43
 
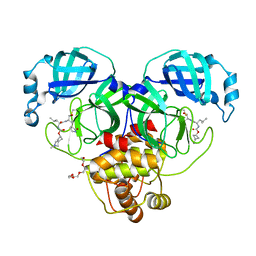 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
4V9S
 
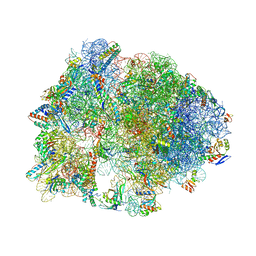 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
1JY6
 
 | |
9BER
 
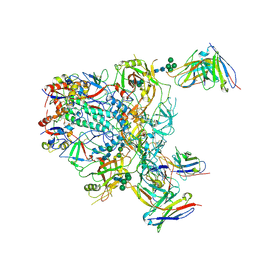 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BKO
 
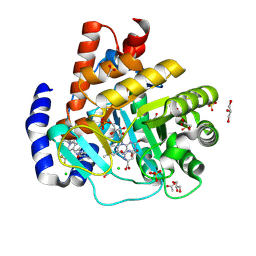 | | DHODH in complex with Ligand 26 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-[(2R)-1,1,1-trifluoropropan-2-yl]isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
1Y9K
 
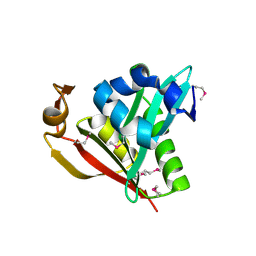 | | IAA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | IAA acetyltransferase | | Authors: | Nocek, B.P, Osipiuk, J, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-15 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A crystal structure of IAA acetyltransferase from Bacillus cereus
To be Published
|
|
1Y9U
 
 | |
7TCZ
 
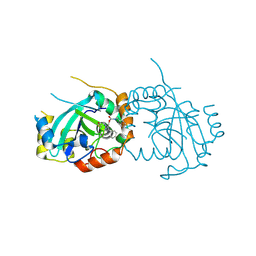 | | Human cytomegalovirus protease mutant (C84A, C87A, C138A, C202A) in complex with inhibitor | | Descriptor: | Assemblin, [1-(2-oxopropyl)-4-phenyl-1H-1,2,3-triazol-5-yl]methyl benzylcarbamate | | Authors: | Hulce, K.R, Bohn, M, Ongpipattanakul, C, Jaishankar, P, Renslo, A.R, Craik, C.S. | | Deposit date: | 2021-12-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibiting a dynamic viral protease by targeting a non-catalytic cysteine.
Cell Chem Biol, 29, 2022
|
|
1JHL
 
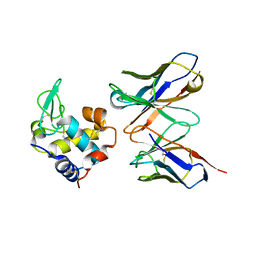 | | THREE-DIMENSIONAL STRUCTURE OF A HETEROCLITIC ANTIGEN-ANTIBODY CROSS-REACTION COMPLEX | | Descriptor: | IGG1-KAPPA D11.15 FV (HEAVY CHAIN), IGG1-KAPPA D11.15 FV (LIGHT CHAIN), PHEASANT EGG WHITE LYSOZYME | | Authors: | Chitarra, V, Alzari, P.M, Bentley, G.A, Bhat, T.N, Eisele, J.-L, Poljak, R.J. | | Deposit date: | 1993-05-04 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4UX8
 
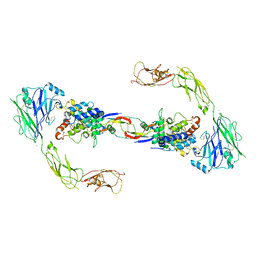 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
1YF2
 
 | | Three-dimensional structure of DNA sequence specificity (S) subunit of a type I restriction-modification enzyme and its functional implications | | Descriptor: | Type I restriction-modification enzyme, S subunit | | Authors: | Kim, J.S, Degiovanni, A, Jancarik, J, Adams, P.D, Yokota, H.A, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-12-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1K62
 
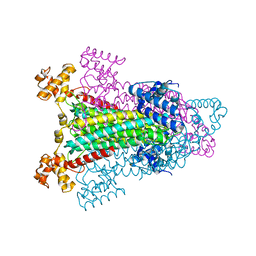 | |
1KAK
 
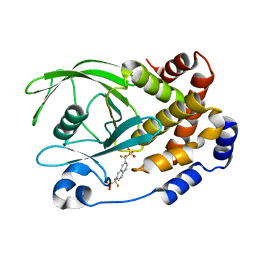 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, {[7-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALEN-2-YL]-DIFLUORO-METHYL}-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
9ARD
 
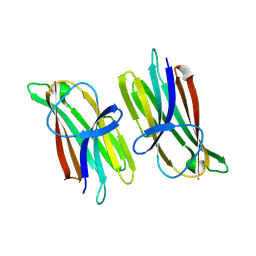 | |
1KB8
 
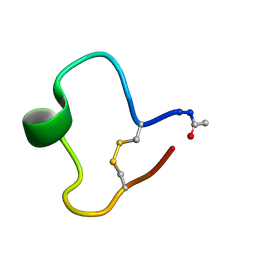 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
