4ERA
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EHL
 
 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P.D, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
4ESB
 
 | | Crystal structure of PadR-like transcriptional regulator (BC4206) from Bacillus cereus strain ATCC 14579 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Fibriansah, G, Kovacs, A.T, Kuipers, O.P, Thunnissen, A.M.W.H. | | Deposit date: | 2012-04-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Two Transcriptional Regulators from Bacillus cereus Define the Conserved Structural Features of a PadR Subfamily.
Plos One, 7, 2012
|
|
4E49
 
 | |
4E6U
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4EHN
 
 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P.D, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
6L3Y
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-C | | Descriptor: | (3R)-3-[[(3S)-3-ethylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Babbar, P, Sharma, A, Mishra, S, Manickam, Y, Harlos, K. | | Deposit date: | 2019-10-15 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
4EIF
 
 | | Crystal structure of cytochrome c6C L50Q mutant from Synechococcus sp. PCC 7002 | | Descriptor: | CHLORIDE ION, Cytochrome c6, HEME C, ... | | Authors: | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
4EO1
 
 | | crystal structure of the TolA binding domain from the filamentous phage IKe | | Descriptor: | Attachment protein G3P, MAGNESIUM ION | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
2PYP
 
 | | PHOTOACTIVE YELLOW PROTEIN, PHOTOSTATIONARY STATE, 50% GROUND STATE, 50% BLEACHED | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Borgstahl, G.E.O, Ng, K, Ren, Z, Pradervand, C, Burke, P, Srajer, V, Teng, T, Schildkamp, W, Mcree, D.E, Moffat, K, Getzoff, E.D. | | Deposit date: | 1997-02-03 | | Release date: | 1998-04-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a protein photocycle intermediate by millisecond time-resolved crystallography.
Science, 275, 1997
|
|
4E6T
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4EQ9
 
 | | 1.4 Angstrom Crystal Structure of ABC Transporter Glutathione-Binding Protein GshT from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Glutathione | | Descriptor: | ABC transporter substrate-binding protein-amino acid transport, GLUTATHIONE | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Light, S.H, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom Crystal Structure of Putative ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Glutathione.
TO BE PUBLISHED
|
|
4E93
 
 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | Authors: | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
8RU2
 
 | | Structure of the F-actin barbed end bound by formin mDia1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8R5T
 
 | |
4EGI
 
 | | Hsp90-alpha ATPase domain in complex with 2-Amino-4-ethylthio-6-methyl-1,3,5-triazine | | Descriptor: | 4-(ethylsulfanyl)-6-methyl-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors.
J Biomol Screen, 17, 2012
|
|
4EHF
 
 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P.D, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
6LYJ
 
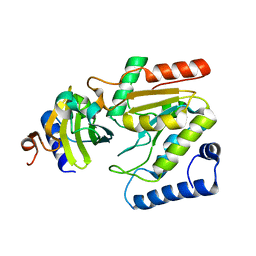 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
6LZ7
 
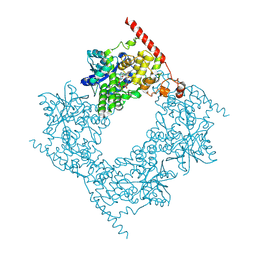 | |
2PXH
 
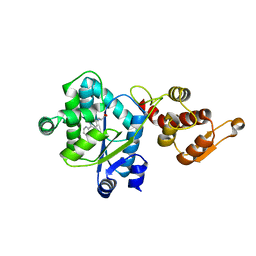 | | Crystal structure of a bipyridylalanyl-tRNA synthetase | | Descriptor: | 3-(2,2'-BIPYRIDIN-5-YL)-L-ALANINE, Tyrosyl-tRNA synthetase | | Authors: | Liu, W, Xie, J, Schultz, P.G. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A genetically encoded bidentate, metal-binding amino acid.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
4EP7
 
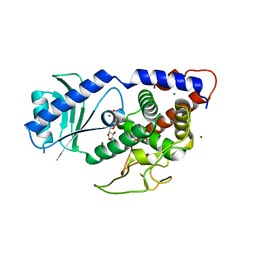 | |
4EPK
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
6LOT
 
 | | Crystal structure of DUSP22 mutant_N128D | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
2POL
 
 | |
2Q7N
 
 | | Crystal structure of Leukemia inhibitory factor in complex with LIF receptor (domains 1-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huyton, T, Zhang, J.G, Nicola, N.A, Garrett, T.P.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
