6Z5M
 
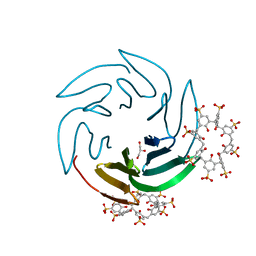 | | The RSL - sulfonato-calix[8]arene complex, I23 form, Gly-HCl pH 2.2 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
5VXB
 
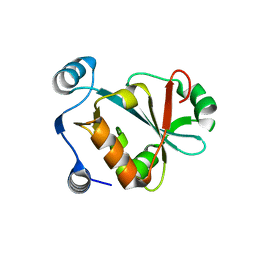 | |
7M6B
 
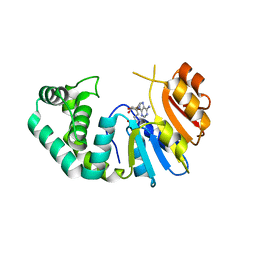 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
5VRT
 
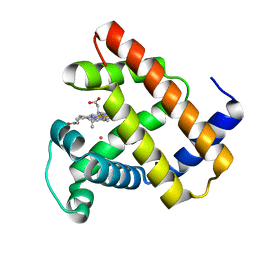 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | Descriptor: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
7RAI
 
 | |
6ZHP
 
 | |
5W26
 
 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 4-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},3~{R},5~{R})-3-acetamido-5-fluoranyl-2-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-05 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7RMF
 
 | | Substrate-bound Ura7 filament at low pH | | Descriptor: | CTP synthase | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
5W2U
 
 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 7-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,4R,5R,6S)-5-acetamido-6-[(2S)-2,3-bis(oxidanyl)propyl]-2,3-bis(fluoranyl)-4-oxidanyl-oxane-2-carboxylic acid, (2~{S},3~{R},4~{R},5~{R})-3-acetamido-2-[(2~{S})-2,3-bis(oxidanyl)propyl]-5-fluoranyl-4-oxidanyl-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
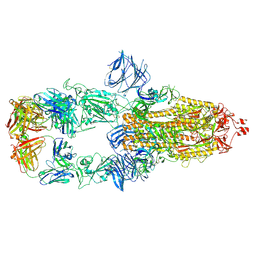 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
5VR3
 
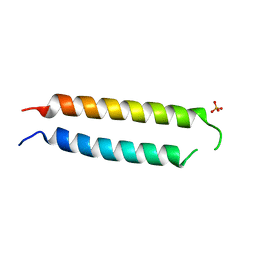 | | Crystal structure of the BRS domain of BRAF | | Descriptor: | BRAF, SULFATE ION | | Authors: | Thevakumaran, N, Maisonneuve, P, Kurinov, I, Lavoie, H, Marullo, S.A, Sahmi, M, Jin, T, Therrien, M, Sicheri, F. | | Deposit date: | 2017-05-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
6YZH
 
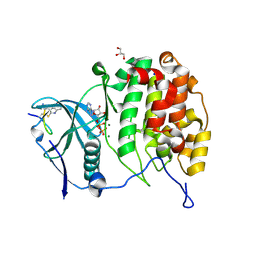 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
5VSL
 
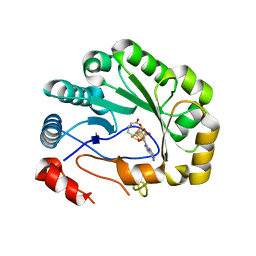 | | Crystal structure of viperin with bound [4Fe-4S] cluster and S-adenosylhomocysteine (SAH) | | Descriptor: | IRON/SULFUR CLUSTER, Radical S-adenosyl methionine domain-containing protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8BAY
 
 | | Crystal Structure of IDH1 variant R132C S280F in complex with NADPH, Ca2+ and 3-butyl-2-oxoglutarate | | Descriptor: | (R)-3-butyl-2-oxopentanedioic acid, (S)-3-butyl-2-oxopentanedioic acid, CALCIUM ION, ... | | Authors: | Rabe, P, Schofield, C.J, Reinbold, R, Brewitz, L. | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2.
J.Biol.Chem., 299, 2023
|
|
7MTT
 
 | |
6Z0Y
 
 | |
6Z0X
 
 | |
6Z0F
 
 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
6Z14
 
 | | Structure of Bifidobacterium bifidum GH20 beta-N-beta-N-acetylhexosaminidase E553Q variant in complex with 4MU-6SGlcNAc-derived oxazoline | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylhexosaminidase, NITRATE ION, ... | | Authors: | He, Y, Jin, Y, Rizkallah, P, Chen, P. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the GH20 beta-N-beta-N-acetylhexosaminidase from Bifidobacterium bifidum
To Be Published
|
|
