4XVT
 
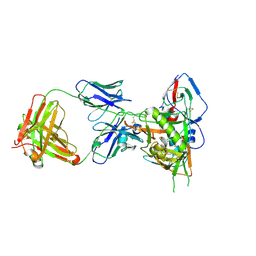 | | Crystal structure of HIV-1 93TH057 coreE gp120 with antibody 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) Light chain, ENVELOPE GLYCOPROTEIN GP120 OF HIV-1 CLADE A/E, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4RCR
 
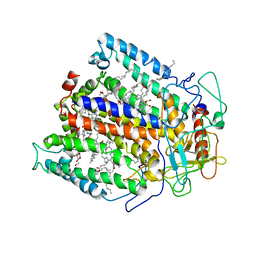 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
5LVJ
 
 | |
1OCI
 
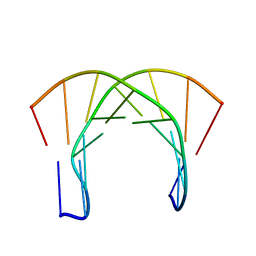 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
5ID7
 
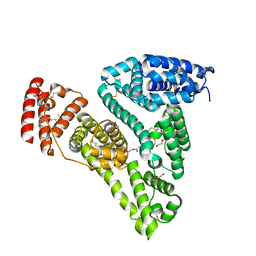 | | Crystal structure of human serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
7ZY3
 
 | | Room temperature structure of Archaerhodopsin-3 obtained 110 ns after photoexcitation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kwan, T.O.C, Judge, P.J, Moraes, I, Watts, A, Axford, D, Bada Juarez, J.F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
5IDP
 
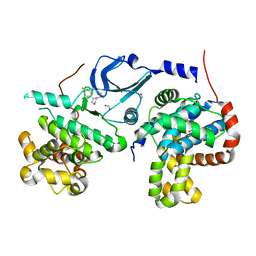 | | CDK8-CYCC IN COMPLEX WITH (3-Amino-1H-indazol-5-yl)-[(S)-2-(4-fluoro-phenyl)-piperidin-1-yl]-methanone | | Descriptor: | (3-amino-1H-indazol-5-yl)[(2S)-2-(4-fluorophenyl)piperidin-1-yl]methanone, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
8IBY
 
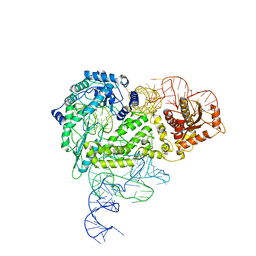 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
1K54
 
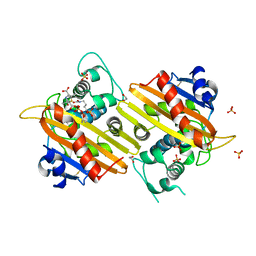 | | OXA-10 class D beta-lactamase partially acylated with reacted 6beta-(1-hydroxy-1-methylethyl) penicillanic acid | | Descriptor: | (1R)-2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Beta lactamase OXA-10, ... | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3ZQT
 
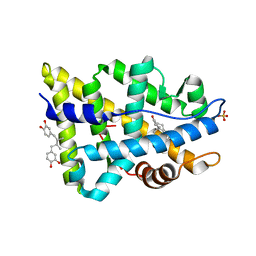 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
3ZRK
 
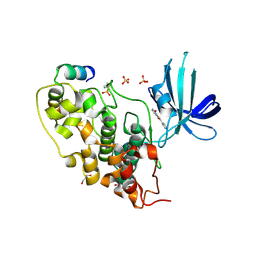 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 2-(4-PYRIDINYL)FURO[3,2-C]PYRIDIN-4(5H)-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6ZMZ
 
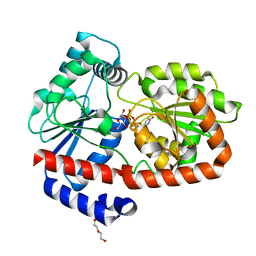 | | UDPG-bound Trehalose transferase from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Trehalose phosphorylase/synthase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
6ZN1
 
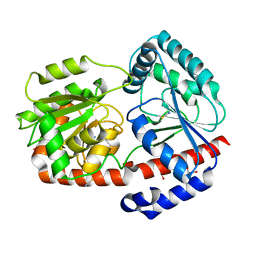 | | Trehalose transferase bound to alpha-D-glucopyranosyl-beta-galactopyranose from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOCYANATE ION, Trehalose phosphorylase/synthase, ... | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
ACS Catal, 10, 2020
|
|
3ZRL
 
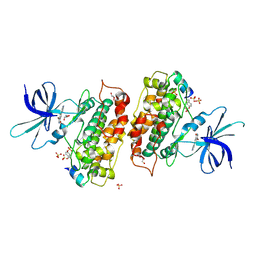 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 7-BROMO-2-PYRIDIN-4-YL-5H-THIENO[3,2-C]PYRIDIN-4-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6C7T
 
 | |
1XZU
 
 | |
2VIJ
 
 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(1,2,3,4- tetrahydro-1-naphthalenylamino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(1S)-1,2,3,4-tetrahydronaphthalen-1-ylamino]propyl}-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Beswick, P, Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Gleave, R, Hawkins, J, Hussain, I, Johnson, C.N, Macpherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Skidmore, J, Soleil, V, Smith, K.J, Stanway, S, Stemp, G, Stuart, A, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-04 | | Release date: | 2008-01-29 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 3: Identification of Hydroxy Ethylamines (Heas) with Nanomolar Potency in Cells.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6CAI
 
 | |
6ZMB
 
 | | Structure of the native tRNA-Monooxygenase enzyme MiaE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
8V4Y
 
 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
7UMJ
 
 | | Crystal structure of recombinant Solieria filiformis lectin (rSfL) | | Descriptor: | Lectin SfL-1, SULFATE ION | | Authors: | Chaves, R.P, Bezerra, E.H.S, da Silva, F.M.S, Carneiro, R.F, Sampaio, A.H, Rocha, B.A.M, Nagano, C.S. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural study and antimicrobial and wound healing effects of lectin from Solieria filiformis (Kutzing) P.W.Gabrielson.
Biochimie, 214, 2023
|
|
6UDB
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | DI(HYDROXYETHYL)ETHER, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ZMC
 
 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 2000 bar using the high pressure freezing method | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGON, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
