8QZ3
 
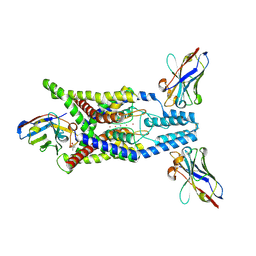 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
4YAC
 
 | | Crystal structure of LigO in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
6TDE
 
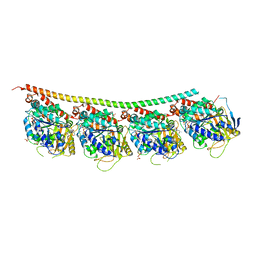 | | Tubulin-inhibitor complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Discovery of dihydrofuranoallocolchicinoids - Highly potent antimitotic agents with low acute toxicity.
Eur.J.Med.Chem., 207, 2020
|
|
6G4Z
 
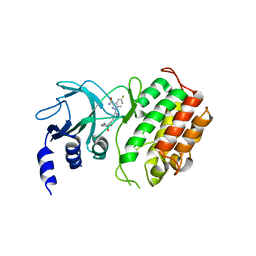 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | | Descriptor: | 5-fluoranyl-1-[4-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrol-3-yl]ethynyl]pyridin-2-yl]indazole-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Leonardo-Silvestre, H, McEwan, P.A, Hymowitz, S.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
5LNW
 
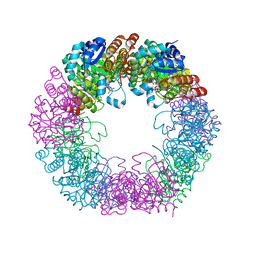 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
3J5L
 
 | | Structure of the E. coli 50S subunit with ErmBL nascent chain | | Descriptor: | 23S ribsomal RNA, 5'-R(*CP*(MA6))-3', 5'-R(*CP*CP*A)-3', ... | | Authors: | Arenz, S, Ramu, H, Gupta, P, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Mankin, A.S, Wilson, D.N. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide.
Nat Commun, 5, 2014
|
|
7BXG
 
 | | MavC-UBE2N-Ub complex | | Descriptor: | MavC, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
8I8N
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution.
To Be Published
|
|
7TN2
 
 | | Composite model of a Chd1-nucleosome complex in the nucleotide-free state derived from 2.3A and 2.7A Cryo-EM maps | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6NE3
 
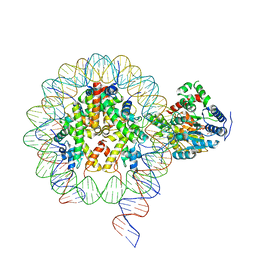 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | Authors: | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | Deposit date: | 2018-12-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
6TVK
 
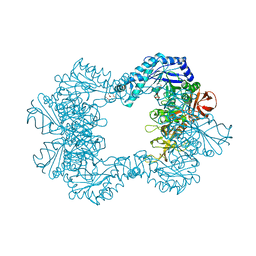 | | Alpha-L-fucosidase isoenzyme 2 from Paenibacillus thiaminolyticus | | Descriptor: | Alpha-L-fucosidase, GLYCEROL, HEXATANTALUM DODECABROMIDE, ... | | Authors: | Kovalova, T, Koval, T, Lipovova, P, Dohnalek, J. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first structure-function study of GH151 alpha-l-fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
Febs J., 2022
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
6TQD
 
 | | D00-D0 domain of Intimin | | Descriptor: | BROMIDE ION, CHLORIDE ION, Intimin, ... | | Authors: | Weikum, J, Leo, J.C, Morth, J.P. | | Deposit date: | 2019-12-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
5LV3
 
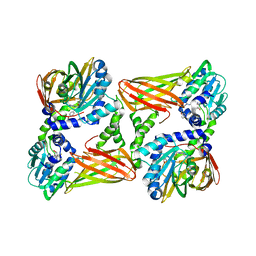 | | Crystal structure of mouse CARM1 in complex with ligand LH1561Br | | Descriptor: | 5-[[2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethylamino]methyl]-4-azanyl-1-[2-(4-bromanylphenoxy)ethyl]pyrimidin-2-one, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
8AV6
 
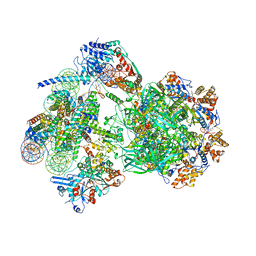 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7QT9
 
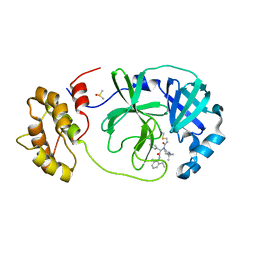 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011584 | | Descriptor: | DIMETHYL SULFOXIDE, N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide, Non-structural protein 6 | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
8GJW
 
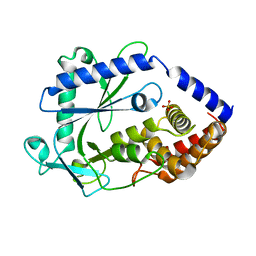 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8IQ5
 
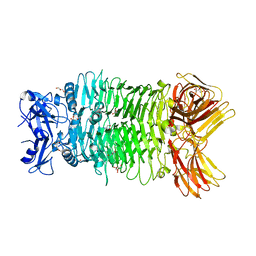 | | Crystal structure of trimeric K2-2 TSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, GLYCEROL, ... | | Authors: | Ye, T.J, Huang, K.F, Ko, T.P. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
7QT6
 
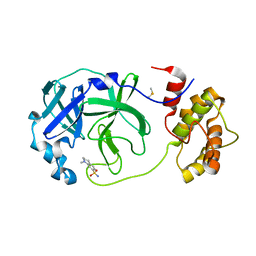 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7QTQ
 
 | | Structure of Native, iodinated bovine thyroglobulin solved on strepavidin affinity grids. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, ... | | Authors: | Marechal, N, Weitz, J.C, Serrano, B.P, Zhang, X. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Formation of thyroid hormone revealed by a cryo-EM structure of native bovine thyroglobulin.
Nat Commun, 13, 2022
|
|
7QT5
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT7
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011520 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6G4Y
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 1a | | Descriptor: | 10-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrolidin-3-yl]ethynyl]-~{N}3-(oxan-4-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Hole, A.J, Hymowitz, S.G, McEwan, P.A. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
