7PEM
 
 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
7NMG
 
 | | Human MHC Class I, A24 Allele presenting LWM, Complex with 4C6 TCR | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diabetes epitope LWMRLLPLL, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Cole, D.K, Wall, A. | | Deposit date: | 2021-02-23 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Human MHC Class I, A24 Allele presenting LWM, Complex with 4C6 TCR
To Be Published
|
|
8FMD
 
 | |
8FME
 
 | |
8FOS
 
 | |
8I8J
 
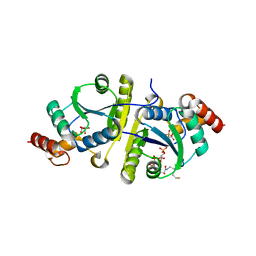 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.07 A resolution.
To Be Published
|
|
6SNH
 
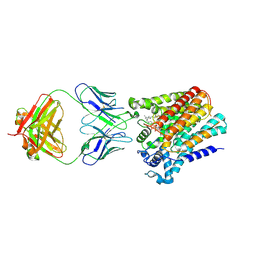 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
8I8K
 
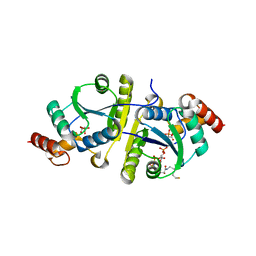 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.13 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.13 A resolution.
To Be Published
|
|
6SSA
 
 | | Human Leukocyte Antigen Class I A02 Carrying LLWNGPMQV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
8I8L
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.23 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.23 A resolution.
To Be Published
|
|
5J43
 
 | |
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
7PI1
 
 | | Bacillus subtilis PabB | | Descriptor: | Aminodeoxychorismate synthase component 1, MAGNESIUM ION, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Crystal structure of Bacillus subtilis PabB, component 1.
To be published
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
5FJW
 
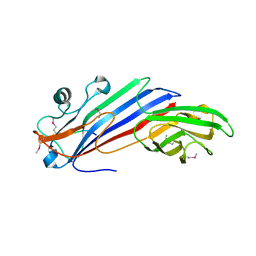 | |
8FQC
 
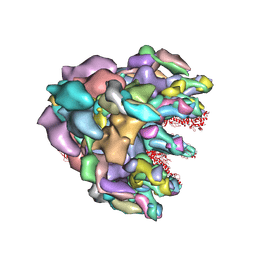 | | Structure of baseplate with receptor binding complex of Agrobacterium phage Milano | | Descriptor: | Baseplate Centerpiece, gp25, Baseplate Central Spike, ... | | Authors: | Sonani, R.R, Leiman, P.G, Wang, F, Kreutzberger, M.A.B, Sebastian, A, Esteves, N.C, Kelly, R.J, Scharf, B, Egelman, E.H. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An extensive disulfide bond network prevents tail contraction in Agrobacterium tumefaciens phage Milano.
Nat Commun, 15, 2024
|
|
5MIT
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (240 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
7ANQ
 
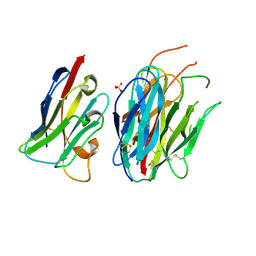 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
4XTS
 
 | | Salmonella typhimurium AhpC T43A mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
3IYQ
 
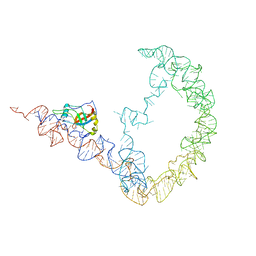 | | tmRNA-SmpB: a journey to the center of the bacterial ribosome | | Descriptor: | SsrA-binding protein, tmRNA | | Authors: | Weis, F, Bron, P, Giudice, E, Rolland, J.P, Thomas, D, Felden, B, Gillet, R. | | Deposit date: | 2010-04-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Embo J., 29, 2010
|
|
3IS8
 
 | | Structure of mineralized Bfrb soaked with FeSO4 from Pseudomonas aeruginosa to 2.25A Resolution | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
5MFB
 
 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
