5XNV
 
 | | Crystal structure of YEATS2 YEATS bound to H3K27ac peptide | | Descriptor: | ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Li, H.T, Guan, H.P, Zhao, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer.
Nat Commun, 8, 2017
|
|
6FM0
 
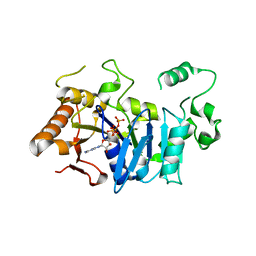 | | Deoxyguanylosuccinate synthase (DgsS) and ATP structure at 1.7 Angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
8CGW
 
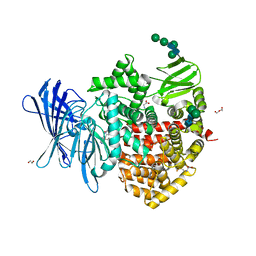 | | Insulin-regulated aminopeptidase (IRAP) in complex with an allosteric benzopyran-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanisms of Allosteric Inhibition of Insulin-Regulated Aminopeptidase.
J.Mol.Biol., 436, 2024
|
|
129D
 
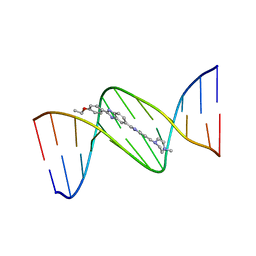 | | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') COMPLEXED WITH HOECHST 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
127D
 
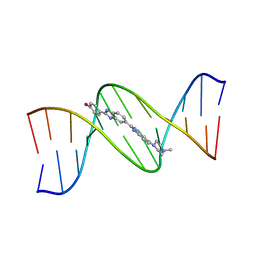 | | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') COMPLEXED WITH HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
7SU9
 
 | |
129L
 
 | |
8C51
 
 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
130L
 
 | |
6FUF
 
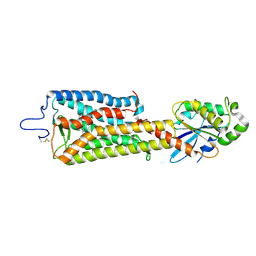 | | Crystal structure of the rhodopsin-mini-Go complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(o) subunit alpha, RETINAL, ... | | Authors: | Tsai, C.-J, Weinert, T, Muehle, J, Pamula, F, Nehme, R, Flock, T, Nogly, P, Edwards, P.C, Carpenter, B, Gruhl, T, Ma, P, Deupi, X, Standfuss, J, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2018-02-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.117 Å) | | Cite: | Crystal structure of rhodopsin in complex with a mini-Gosheds light on the principles of G protein selectivity.
Sci Adv, 4, 2018
|
|
131L
 
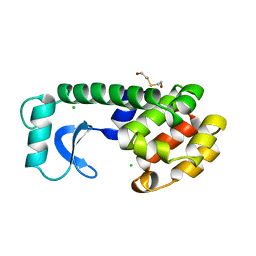 | |
7T9X
 
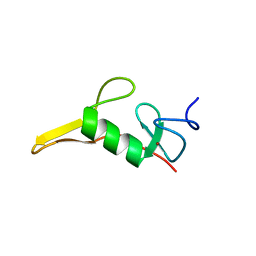 | | Saccharomyces cerevisiae Pex12 RING domain | | Descriptor: | Peroxisome assembly protein 12, ZINC ION | | Authors: | Feng, P, Rapoport, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel.
Nature, 607, 2022
|
|
7O9Q
 
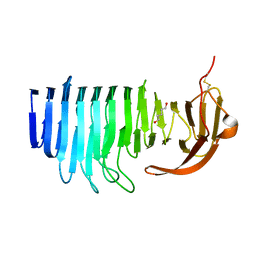 | |
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
5JQR
 
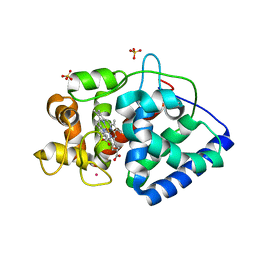 | | The Structure of Ascorbate Peroxidase Compound II formed by reaction with m-CPBA | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kwon, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2016-05-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Nat Commun, 7, 2016
|
|
7O9P
 
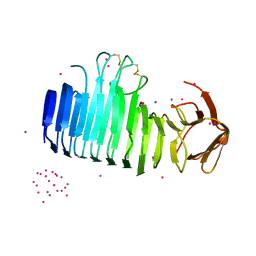 | |
7PEW
 
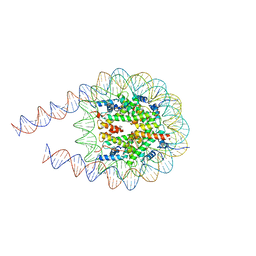 | |
7PEX
 
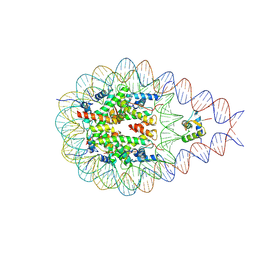 | |
5MMS
 
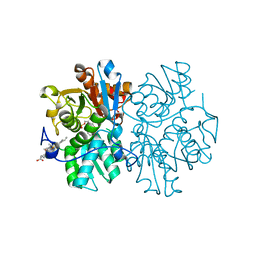 | | Human cystathionine beta-synthase (CBS) p.P49L delta409-551 variant | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Vicente, J.B, Colaco, H.G, Malagrino, F, Santo, P.E, Gutierres, A, Bandeiras, T.M, Leandro, P, Brito, J.A, Giuffre, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Clinically Relevant Variant of the Human Hydrogen Sulfide-Synthesizing Enzyme Cystathionine beta-Synthase: Increased CO Reactivity as a Novel Molecular Mechanism of Pathogenicity?
Oxid Med Cell Longev, 2017, 2017
|
|
7PF5
 
 | |
1AI0
 
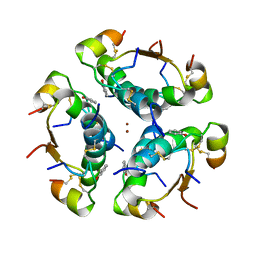 | | R6 HUMAN INSULIN HEXAMER (NON-SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
7PF6
 
 | |
6G7I
 
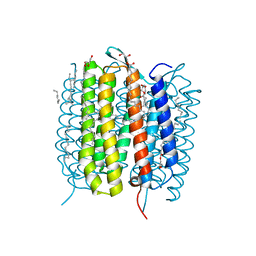 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 49-406 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
7PFD
 
 | |
