5V53
 
 | |
2AES
 
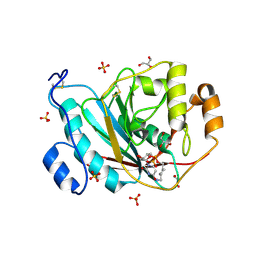 | | Crystal Structure of Human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4Gal-T1) in Complex with GlcNAc-beta1,2-Man-alpha1,3-Man-beta-OR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
1D5W
 
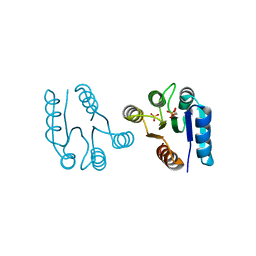 | | PHOSPHORYLATED FIXJ RECEIVER DOMAIN | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Birck, C, Mourey, L, Gouet, P, Fabry, B, Schumacher, J, Rousseau, P, Kahn, D, Samama, J.P. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes induced by phosphorylation of the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1IJG
 
 | | Structure of the Bacteriophage phi29 Head-Tail Connector Protein | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4GQM
 
 | |
4B30
 
 | |
1SJL
 
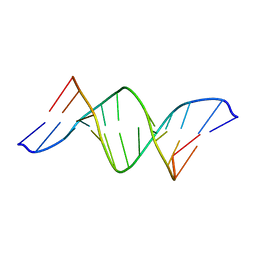 | | A DUPLEX DNA WITH AN ABASIC SITE IN A DA TRACT, BETA FORM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*TP*(AAB)P*TP*TP*GP*CP*G)-3') | | Authors: | Wang, K.Y, Parker, S.A, Goljer, I, Bolton, P.H. | | Deposit date: | 1997-07-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a duplex DNA with an abasic site in a dA tract.
Biochemistry, 36, 1997
|
|
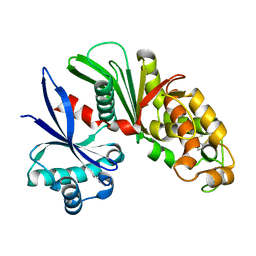
3AAP
 
 | |
4HGA
 
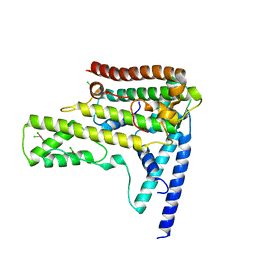 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1NJQ
 
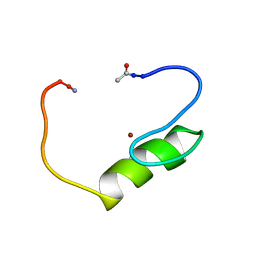 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
2PWS
 
 | | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 A resolution | | Descriptor: | IBUPROFEN, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-13 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 resolution
To be Published
|
|
1NI2
 
 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
1NIJ
 
 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
1SSC
 
 | | THE 1.6 ANGSTROMS STRUCTURE OF A SEMISYNTHETIC RIBONUCLEASE CRYSTALLIZED FROM AQUEOUS ETHANOL. COMPARISON WITH CRYSTALS FROM SALT SOLUTIONS AND WITH RNASE A FROM AQUEOUS ALCOHOL SOLUTIONS | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE A | | Authors: | De Mel, V.S.J, Doscher, M.S, Martin, P.D, Rodier, F, Edwards, B.F.P. | | Deposit date: | 1994-10-05 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
4N2L
 
 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
3PKJ
 
 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
1YWG
 
 | | The structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Satchell, J.F, Klonis, N, Malby, R.L, Adisa, A, Alpyurek, A.E, Colman, P.M, Tilley, L. | | Deposit date: | 2005-02-17 | | Release date: | 2005-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4H69
 
 | |
1NAB
 
 | | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', 7-[5-(4-AMINO-5-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY)-4-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY]-6,9,11-TRIHYDROXY-9-(2-HYDROXY-ACETYL)-7,8,9,10-TETRAHYDRO-NAPHTHACENE-5,12-DIONE | | Authors: | Temperini, C, Messori, L, Orioli, P, Di Bugno, C, Animati, F, Ughetto, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes
Nucleic Acids Res., 31, 2003
|
|
4BC6
 
 | | Crystal structure of human serine threonine kinase-10 bound to novel Bosutinib Isoform 1, previously thought to be Bosutinib | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Vollmar, M, Szklarz, M, Chaikuad, A, Elkins, J, Savitsky, P, Azeez, K.A, Salah, E, Krojer, T, Canning, P, Muniz, J.R.C, Bountra, C, Arrowsmith, C.H, von Delft, F, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2012-10-01 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Serine Threonine Kinase- 10 Bound to Novel Bosutinib Isoform 1, Previously Thought to be Bosutinib
To be Published
|
|
4N7K
 
 | | Zinc Substituted Reaction Center of the Rhodobacter sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, FE (III) ION, ... | | Authors: | Hardjasa, A, Murphy, M.E.P. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors.
Biochim.Biophys.Acta, 1837, 2013
|
|
4B15
 
 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
2PWA
 
 | | Crystal Structure of the complex of Proteinase K with Alanine Boronic acid at 0.83A resolution | | Descriptor: | ALANINE BORONIC ACID, CALCIUM ION, NITRATE ION, ... | | Authors: | Jain, R, Singh, N, Perbandt, M, Betzel, C, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Crystal structure of the complex of Proteinase K with Alanine Boronic Acid at 0.83A Resolution
To be Published
|
|
2XSG
 
 | | Structure of the gh92 family glycosyl hydrolase ccman5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CCMAN5, ... | | Authors: | Baranova, E, Tiels, P, Remaut, H. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Glycosidase Enables Mannose-6-Phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes.
Nat.Biotechnol., 30, 2012
|
|
4JBG
 
 | | 1.75A resolution structure of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
