5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WE5
 
 | | The crystal structure of hemagglutinin from A/Port Chalmers/1/1973 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4WEG
 
 | | influenza virus neuraminidase N9 in complex 2,3-difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
4WE8
 
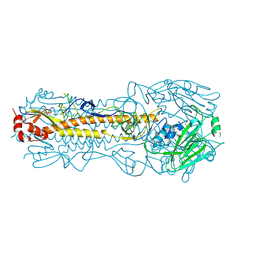 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
6E0M
 
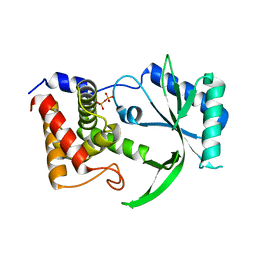 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase | | Descriptor: | DIPHOSPHATE, cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
5I9L
 
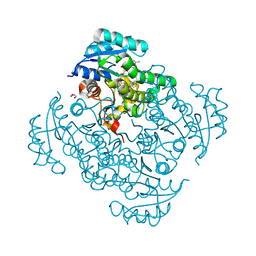 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | Descriptor: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
6E0N
 
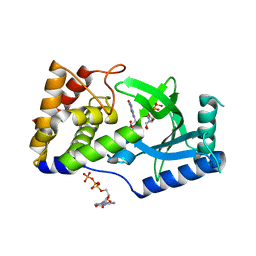 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0L
 
 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase with Apcpp and Upnpp | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
1AGU
 
 | | THE SOLUTION NMR STRUCTURE OF THE C10R ADDUCT OF BENZO[A]PYRENE-DIOL-EPOXIDE AT THE N6 POSITION OF ADENINE OF AN 11 BASE-PAIR OLIGONUCLEOTIDE SEQUENCE CODING FOR AMINO ACIDS 60-62 OF THE PRODUCT OF THE N-RAS PROTOONCOGENE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*CP*EP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Zegar, I.S, Stone, M.P. | | Deposit date: | 1997-03-25 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Adduction of the human N-ras codon 61 sequence with (-)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a] pyrene: structural refinement of the intercalated SRSR(61,2) (-)-(7S,8R,9S,1 0R)-N6-[10-(7,8,9,10- tetrahydrobenzo[a]pyrenyl)]-2'-deoxyadenosyl adduct from 1H NMR
Biochemistry, 35, 1996
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | Descriptor: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E2P
 
 | |
6EBE
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EGS
 
 | | Crystal structure of the GalNAc-T2 F104S mutant in complex with UDP-GalNAc | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | de las Rivas, M, Coelho, H, Diniz, A, Lira-Navarrete, E, Jimenez-Barbero, J, Schjoldager, K.T, Bennett, E.P, Vakhrushev, S.Y, Clausen, H, Corzana, F, Marcelo, F, Hurtado-Guerrero, R. | | Deposit date: | 2017-09-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analysis of a GalNAc-T2 Mutant Reveals an Induced-Fit Catalytic Mechanism for GalNAc-Ts.
Chemistry, 24, 2018
|
|
6EJJ
 
 | | Structure of a glycosyltransferase / state 2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, NerylNeryl pyrophosphate, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6EK6
 
 | | Crystal structure of KDM5B in complex with S49195a. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Srikannathasan, V, Szykowska, A, Newman, J.A, Ruda, G.F, Strain-Damerell, C, Burgess-Brown, N.A, Vazquez-Rodriguez, S, Wright, M, Brennan, P.E, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of KDM5B in complex with S49195a.
To be published
|
|
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EAS
 
 | | Co-crystal of pseudokinase DRIK1 (drought responsive inactive kinase 1) bound to ENMD-2076 | | Descriptor: | 6-(4-methylpiperazin-1-yl)-N-(5-methyl-1H-pyrazol-3-yl)-2-[(E)-2-phenylethenyl]pyrimidin-4-amine, GLYCEROL, drought responsive inactive kinase 1 | | Authors: | Aquino, B, Counago, R.M, Fala, A.M, Massirer, K.B, Elkins, J.M, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal of pseudokinase DRIK1 with ENMD-2076
To be Published
|
|
1A35
 
 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1ADZ
 
 | |
4UCB
 
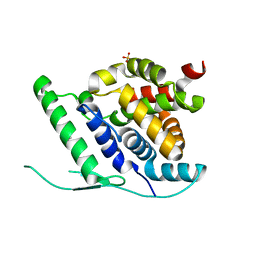 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal peptide of the Phosphoprotein | | Descriptor: | NUCLEOPROTEIN, PHOSPHOSPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
1BA3
 
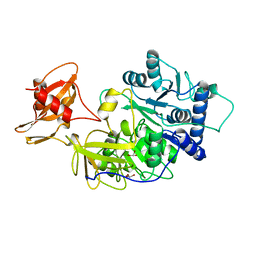 | | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM | | Descriptor: | LUCIFERASE, TRIBROMOMETHANE | | Authors: | Franks, N.P, Jenkins, A, Conti, E, Lieb, W.R, Brick, P. | | Deposit date: | 1998-04-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the inhibition of firefly luciferase by a general anesthetic.
Biophys.J., 75, 1998
|
|
1AP1
 
 | |
