2OKS
 
 | | X-ray Structure of a DNA Repair Substrate Containing an Alkyl Interstrand Crosslink at 1.65 Resolution | | Descriptor: | 5'-D(*CP*CP*AP*AP*(C34)P*GP*TP*TP*GP*G)-3', CALCIUM ION | | Authors: | Swenson, M.C, Paranawithana, S.R, Miller, P.S, Kielkopf, C.L. | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a DNA repair substrate containing an alkyl interstrand cross-link at 1.65 a resolution.
Biochemistry, 46, 2007
|
|
2XHB
 
 | | Crystal structure of DNA polymerase from Thermococcus gorgonarius in complex with hypoxanthine-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', DNA POLYMERASE, HYPOXANTHINE-CONTAINING DNA, ... | | Authors: | Killelea, T, Ghosh, S, Tan, S.S, Heslop, P, Firbank, S.J, Kool, E.T, Connolly, B.A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Probing the Interaction of Archaeal DNA Polymerases with Deaminated Bases Using X-Ray Crystallography and Non-Hydrogen Bonding Isosteric Base Analogues.
Biochemistry, 49, 2010
|
|
1Q9W
 
 | | S45-18 Fab pentasaccharide bisphosphate complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Nguyen, H.P, Seto, N.O, MacKenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2003-08-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
1M10
 
 | | Crystal structure of the complex of Glycoprotein Ib alpha and the von Willebrand Factor A1 Domain | | Descriptor: | Glycoprotein Ib alpha, von Willebrand Factor | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
5W6F
 
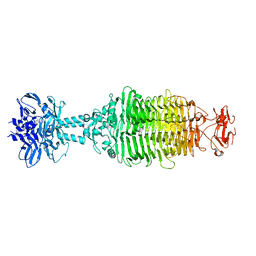 | |
5VYR
 
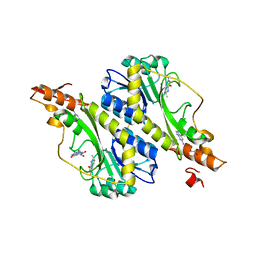 | | Crystal structure of the WbkC formyl transferase from Brucella melitensis | | Descriptor: | (6R)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
2XNZ
 
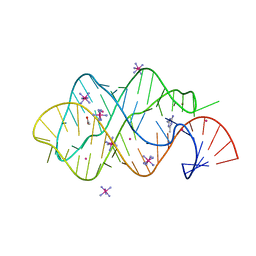 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to acetoguanamine identified by virtual screening | | Descriptor: | 6-METHYL-1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
3LD0
 
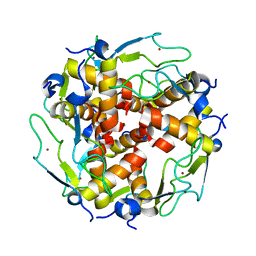 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
4F1N
 
 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
1LY8
 
 | | The crystal structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability and insight into modelling of protein structures | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Houborg, K, Harris, P, Poulsen, J.-C.N, Svendsen, A, Schneider, P, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4F2J
 
 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
2VAI
 
 | | Solution structure of a B-DNA hairpin at high pressure | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural change in a B-DNA helix with hydrostatic pressure.
Nucleic Acids Res., 36, 2008
|
|
4S25
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S2A
 
 | | Crystal structure of Caulobacter crescentus ThiC with Fe4S4 cluster at remote site (holo form) | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, Phosphomethylpyrimidine synthase | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S2R
 
 | | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: a cytosolic enzyme with a di-nuclear active site | | Descriptor: | Protein APP-1, ZINC ION | | Authors: | Iyer, S, La-Borde, P, Payne, K.A.P, Parsons, M.R, Turner, A.J, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2015-01-22 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Crystal structure of X-prolyl aminopeptidase from Caenorhabditis elegans: A cytosolic enzyme with a di-nuclear active site.
FEBS Open Bio, 5, 2015
|
|
3LMJ
 
 | | Structure of human anti HIV 21c Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Heavy chain of anti HIV Fab from human 21c antibody, Light chain of anti HIV Fab from human 21c antibody | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-01-30 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1TCO
 
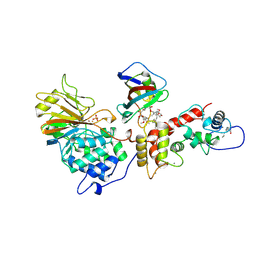 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JWR
 
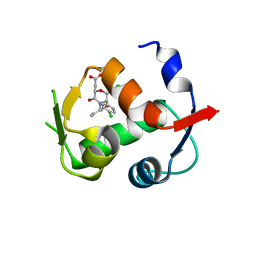 | |
1MET
 
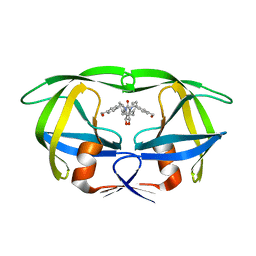 | | HIV-1 MUTANT (V82F) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1TI7
 
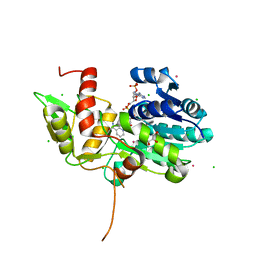 | | CRYSTAL STRUCTURE OF NMRA, A NEGATIVE TRANSCRIPTIONAL REGULATOR, IN COMPLEX WITH NADP AT 1.7A RESOLUTION | | Descriptor: | CHLORIDE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, H.K, Leslie, K, Dodds, A.L, Nutley, M, Cooper, A, Johnson, C, Thompson, P, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
J.Biol.Chem., 278, 2003
|
|
2F73
 
 | | Crystal structure of human fatty acid binding protein 1 (FABP1) | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Kursula, P, Thorsell, A.G, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, van den Berg, S, Weigelt, J, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human FABP1
To be Published
|
|
1SZL
 
 | | F-spondin TSR domain 1 | | Descriptor: | F-spondin | | Authors: | Tossavainen, H, Paakkonen, K, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
1OTO
 
 | | Calcium-binding mutant of the internalin B LRR domain | | Descriptor: | CALCIUM ION, Internalin B | | Authors: | Marino, M, Copp, J, Dramsi, S, Chapman, T, van der Geer, P, Cossart, P, Ghosh, P. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization of the calcium-binding sites of Listeria monocytogenes InlB
Biochem.Biophys.Res.Commun., 316, 2004
|
|
2HM3
 
 | | Nematocyst outer wall antigen, cysteine rich domain NW1 | | Descriptor: | Nematocyst outer wall antigen | | Authors: | Meier, S, Jensen, P.R, Grzesiek, S, Oezbek, S. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Sequence-Structure and Structure-Function Analysis in Cysteine-rich Domains Forming the Ultrastable Nematocyst Wall.
J.Mol.Biol., 368, 2007
|
|
