2WDD
 
 | | Termus thermophilus Sulfate thiohydrolase SoxB in complex with Sulfate | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sauve, V, Roversi, P, Leath, K.J, Garman, E.F, Antrobus, R, Lea, S.M, Berks, B.C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the Hydrolysis of a Sulfur-Sulfur Bond Based on the Crystal Structure of the Thiosulfohydrolase Soxb.
J.Biol.Chem., 284, 2009
|
|
2VSG
 
 | | A Structural Motif in the Variant Surface Glycoproteins of Trypanosoma Brucei | | Descriptor: | VARIANT SURFACE GLYCOPROTEIN ILTAT 1.24 | | Authors: | Blum, M.L, Down, J.A, Metcalf, P, Freymann, D.M, Wiley, D.C. | | Deposit date: | 1998-11-19 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural motif in the variant surface glycoproteins of Trypanosoma brucei.
Nature, 362, 1993
|
|
2W8X
 
 | | Structure of the tick ion-channel modulator Ra-KLP | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Paesen, G.C, Siebold, C, Dallas, M, Peers, C, Harlos, K, Nuttall, P.A, Nunn, M.A, Stuart, D.I, Esnouf, R.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-05 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Channel Modulator from the Saliva of the Brown Ear Tick Has a Highly Modified Kunitz/Bpti Structure.
J.Mol.Biol., 389, 2009
|
|
2VU0
 
 | |
6TNK
 
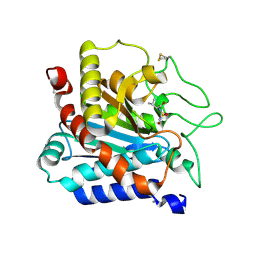 | |
5EKU
 
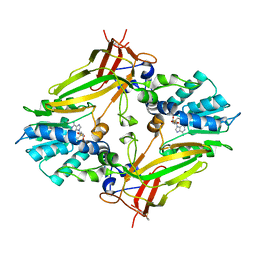 | |
5EOG
 
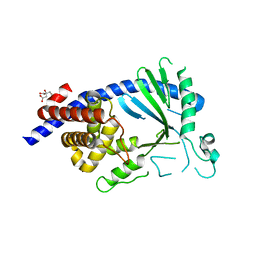 | |
5EGT
 
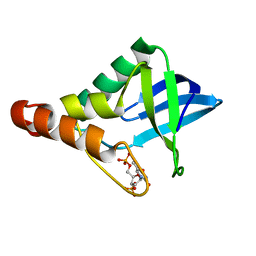 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Siegler, M.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66E at cryogenic temperature
To be Published
|
|
2W20
 
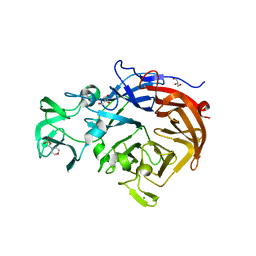 | | Structure of the catalytic domain of the native NanA sialidase from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Xu, G, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-10-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VNZ
 
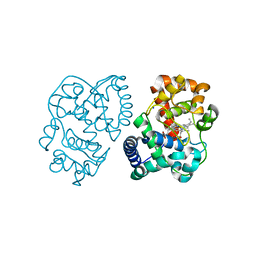 | | Crystal structure of dithinonite reduced soybean ascorbate peroxidase mutant W41A. | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
5EGJ
 
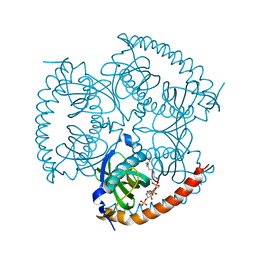 | |
2W86
 
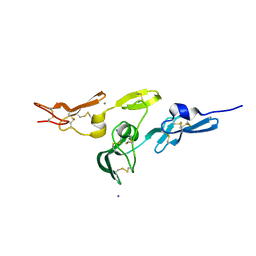 | | Crystal structure of fibrillin-1 domains cbEGF9hyb2cbEGF10, calcium saturated form | | Descriptor: | CALCIUM ION, FIBRILLIN-1, IODIDE ION | | Authors: | Jensen, S.A, Iqbal, S, Lowe, E.D, Redfield, C, Handford, P.A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Interdomain Interactions of a Hybrid Domain: A Disulphide-Rich Module of the Fibrillin/Ltbp Superfamily of Matrix Proteins.
Structure, 17, 2009
|
|
6T9I
 
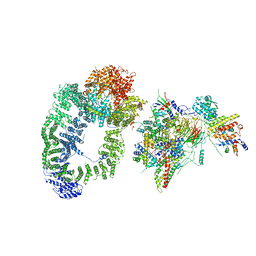 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
2VOG
 
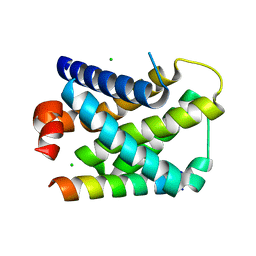 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VWK
 
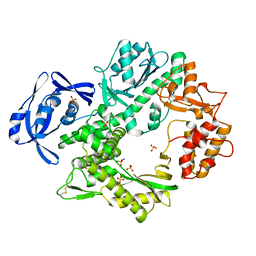 | | Uracil Recognition in Archaeal DNA Polymerases Captured by X-ray Crystallography. V93Q polymerase variant | | Descriptor: | DNA POLYMERASE, SODIUM ION, SULFATE ION | | Authors: | Firbank, S.J, Wardle, J, Heslop, P, Lewis, R.J, Connolly, B.A. | | Deposit date: | 2008-06-26 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uracil Recognition in Archaeal DNA Polymerases Captured by X-Ray Crystallography.
J.Mol.Biol., 381, 2008
|
|
2WB7
 
 | | pT26-6p | | Descriptor: | PT26-6P | | Authors: | Keller, J, Leulliot, N, Soler, N, Collinet, B, Vincentelli, R, Forterre, P, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Protein Encoded by a New Family of Mobile Elements from Euryarchaea Exhibits Three Domains with Novel Folds.
Protein Sci., 18, 2009
|
|
5EP2
 
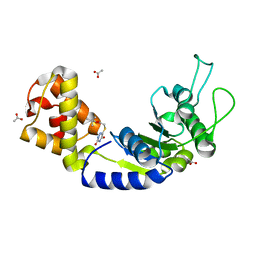 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain in Complex with AzaU Inhibitor | | Descriptor: | 2,2-dimethylpropyl 2-[[3,5-bis(oxidanylidene)-2~{H}-1,2,4-triazin-6-yl]sulfanyl]ethanoate, ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
2VS3
 
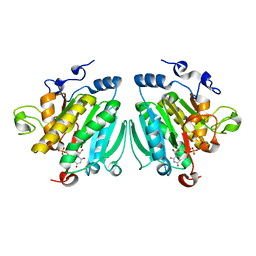 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
4V69
 
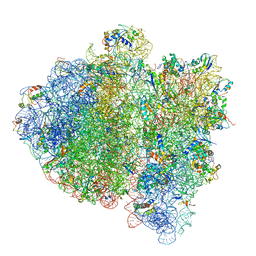 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W5Z
 
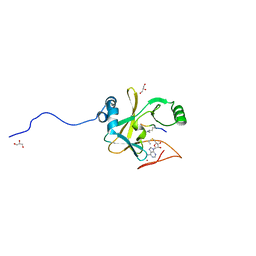 | | Ternary Complex of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and histone peptide. | | Descriptor: | GLYCEROL, HISTONE PEPTIDE, HISTONE-LYSINE N-METHYLTRANSFERASE HRX, ... | | Authors: | Southall, S.M, Wong, P.S, Odho, Z, Roe, S.M, Wilson, J.R. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Mol.Cell, 33, 2009
|
|
2W9Y
 
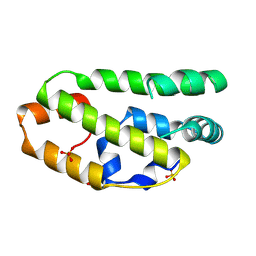 | | The structure of the lipid binding protein Ce-FAR-7 from Caenorhabditis elegans | | Descriptor: | FATTY ACID/RETINOL BINDING PROTEIN PROTEIN 7, ISOFORM A, CONFIRMED BY TRANSCRIPT EVIDENCE, ... | | Authors: | Jordanova, R, Groves, M.R, Tucker, P.A. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-20 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
J.Biol.Chem., 284, 2009
|
|
6T2T
 
 | | Crystal structure of Drosophila melanogaster glutathione S-transferase epsilon 14 in complex with glutathione and 2-methyl-2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Skerlova, J, Lindstrom, H, Sjodin, B, Gonis, E, Neiers, F, Mannervik, B, Stenmark, P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and steroid isomerase activity of Drosophila glutathione transferase E14 essential for ecdysteroid biosynthesis.
Febs Lett., 594, 2020
|
|
2WAG
 
 | | The Structure of a family 25 Glycosyl hydrolase from Bacillus anthracis. | | Descriptor: | GLYCEROL, LYSOZYME, PUTATIVE, ... | | Authors: | Martinez-Fleites, C, Korczynska, J.E, Cope, M, Turkenburg, J.P, Taylor, E.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of a Family Gh25 Lysozyme from Bacillus Anthracis Implies a Neighboring-Group Catalytic Mechanism with Retention of Anomeric Configuration
Carbohydr.Res., 344, 2009
|
|
5EFY
 
 | | Apo-form of SCO3201 | | Descriptor: | Putative tetR-family transcriptional regulator | | Authors: | Waack, P, Hinrichs, W. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 2022
|
|
5EGL
 
 | | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus in complex with Butyryl Coenzyme A, Coenzyme A, and Coenzyme A disulfide | | Descriptor: | Acyl CoA Hydrolase, Butyryl Coenzyme A, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P.S, Forwood, J.K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus
To Be Published
|
|
