1Q5I
 
 | | Crystal structure of bacteriorhodopsin mutant P186A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3VA1
 
 | | Crystal structure of the mammalian MDC1 FHA domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | Authors: | Wu, H.H, Wu, P.Y, Huang, K.F, Kao, Y.Y, Tsai, M.D. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Delineation of MDC1-FHA Domain Binding with CHK2-pThr68.
Biochemistry, 2012
|
|
2N25
 
 | | Solution structure of Miz-1 zinc finger 2 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Lavigne, P. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 13th C2H2 Zinc Finger of Miz-1.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5EMZ
 
 | |
7KI1
 
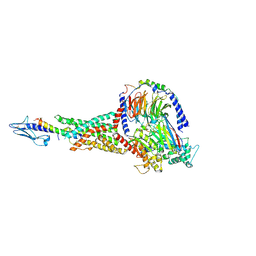 | | Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
1PXH
 
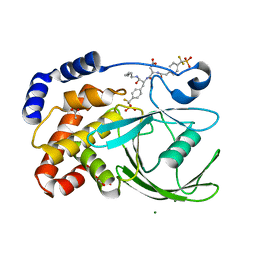 | | Crystal structure of protein tyrosine phosphatase 1B with potent and selective bidentate inhibitor compound 2 | | Descriptor: | ACETIC ACID, MAGNESIUM ION, N-{1-[5-(1-CARBAMOYL-2-MERCAPTO-ETHYLCARBAMOYL)-PENTYLCARBAMOYL]-2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ETHYL}-3-{2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ACETYLAMINO}-SUCCINAMIC ACID, ... | | Authors: | Sun, J.P, Fedorov, A, Lee, S.Y, Guo, X.L, Shen, K, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2003-07-04 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of PTP1B complexed with a potent and selective bidentate inhibitor.
J.Biol.Chem., 278, 2003
|
|
1PXS
 
 | | Structure of Met56Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
6DM7
 
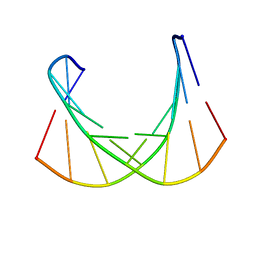 | |
5L2R
 
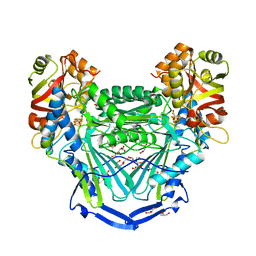 | | Crystal structure of fumarate hydratase from Leishmania major | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DI(HYDROXYETHYL)ETHER, Fumarate hydratase, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7T2Q
 
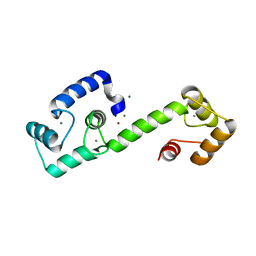 | | PEGylated Calmodulin-1 (K148U) | | Descriptor: | CALCIUM ION, Calmodulin-1, MAGNESIUM ION, ... | | Authors: | Mackay, J.P, Payne, R.J, Patel, K, Dowman, L.J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Site-selective photocatalytic functionalization of peptides and proteins at selenocysteine.
Nat Commun, 13, 2022
|
|
2ZKM
 
 | | Crystal Structure of Phospholipase C Beta 2 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-2, CALCIUM ION | | Authors: | Hicks, S.N, Jezyk, M.R, Gershberg, S, Seifert, J.P, Harden, T.K, Sondek, J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | General and versatile autoinhibition of PLC isozymes
Mol.Cell, 31, 2008
|
|
2ZO9
 
 | | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal ion preference | | Descriptor: | FE (II) ION, MALONATE ION, Phosphohydrolase | | Authors: | Jackson, C.J, Carr, P.D, Ollis, D.L. | | Deposit date: | 2008-05-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1Q1H
 
 | |
3V9E
 
 | | Structure of the L499M mutant of the laccase from B.aclada | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-12-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2MQC
 
 | |
2ZRK
 
 | | MsRecA Q196A dATP form IV | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3S51
 
 | | Structure of FANCI | | Descriptor: | Fanconi anemia group I protein homolog | | Authors: | Pavletich, N.P. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway.
Science, 333, 2011
|
|
3VZN
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VB5
 
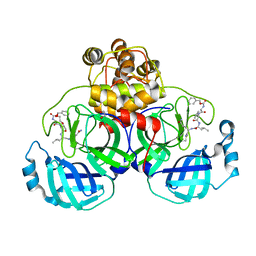 | | Crystal structure of SARS-CoV 3C-like protease with C4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C4Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
6WHB
 
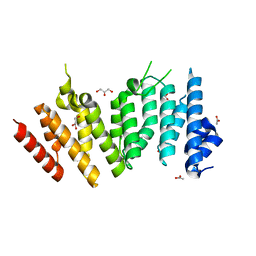 | | MEKK1 TOG domain (548-867) | | Descriptor: | ACETATE ION, GLYCEROL, Mitogen-activated protein kinase kinase kinase 1 | | Authors: | Filipcik, P, Mace, P.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.90032053 Å) | | Cite: | A cryptic tubulin-binding domain links MEKK1 to curved tubulin protomers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2ZRE
 
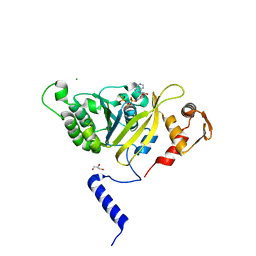 | | MsRecA Q196N ATPgS form IV | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3S3Q
 
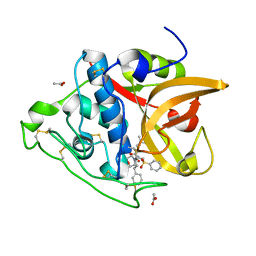 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11017 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
2ZRM
 
 | | MsRecA dATP form IV | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1PET
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1ACW
 
 | | SOLUTION NMR STRUCTURE OF P01, A NATURAL SCORPION PEPTIDE STRUCTURALLY ANALOGOUS TO SCORPION TOXINS SPECIFIC FOR APAMIN-SENSITIVE POTASSIUM CHANNEL, 25 STRUCTURES | | Descriptor: | NATURAL SCORPION PEPTIDE P01 | | Authors: | Blanc, E, Fremont, V, Sizun, P, Meunier, S, Van Rietschoten, J, Thevand, A, Bernassau, J.M, Darbon, H. | | Deposit date: | 1997-02-10 | | Release date: | 1997-04-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P01, a natural scorpion peptide structurally analogous to scorpion toxins specific for apamin-sensitive potassium channel.
Proteins, 24, 1996
|
|
