1MZA
 
 | | crystal structure of human pro-granzyme K | | Descriptor: | pro-granzyme K | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Wilharm, E, Fuentes-Prior, P, Klinkert, W, Bode, W, Jenne, D.E. | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The 2.2-A Crystal Structure of Human Pro-granzyme K Reveals a Rigid Zymogen with Unusual Features
J.BIOL.CHEM., 277, 2002
|
|
4FOQ
 
 | |
3UNG
 
 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Cmr2dHD, ... | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
7R65
 
 | |
4FP3
 
 | |
7RTB
 
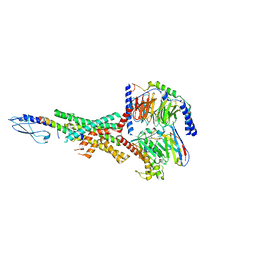 | | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Johnson, R.M, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2021-08-12 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
3HK3
 
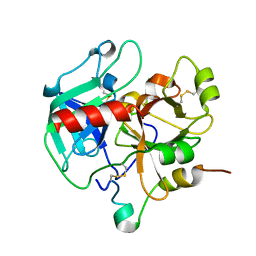 | | Crystal structure of murine thrombin mutant W215A/E217A (one molecule in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
4FPJ
 
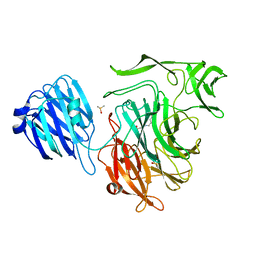 | |
4FPY
 
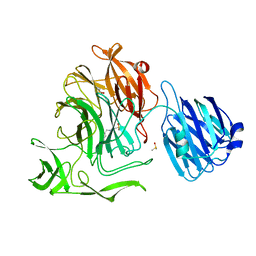 | |
3UO6
 
 | | Aurora A in complex with YL5-083 | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(2-chlorophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8002 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
5KL9
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, COENZYME A, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with CoA
To Be Published
|
|
3UOJ
 
 | | Aurora A in complex with RPM1715 | | Descriptor: | 4-({4-[(2-cyanophenyl)amino]pyrimidin-2-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9003 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
1N4V
 
 | |
3HKJ
 
 | | Crystal structure of human thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
7RPJ
 
 | | Cryo-EM structure of murine Dispatched NNN mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
3HKB
 
 | | Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HRF
 
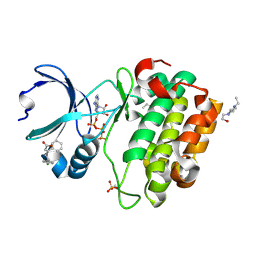 | | Crystal structure of Human PDK1 kinase domain in complex with an allosteric activator bound to the PIF-pocket | | Descriptor: | (2Z)-5-(4-chlorophenyl)-3-phenylpent-2-enoic acid, 3-phosphoinositide-dependent protein kinase 1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hindie, V, Alzari, P.M, Biondi, R.M. | | Deposit date: | 2009-06-09 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1.
Nat.Chem.Biol., 5, 2009
|
|
5KSF
 
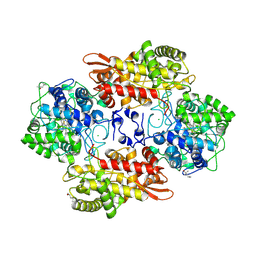 | |
6KVN
 
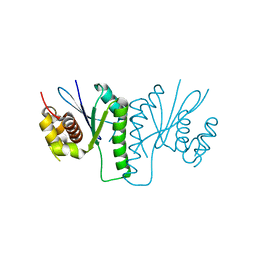 | |
6KZS
 
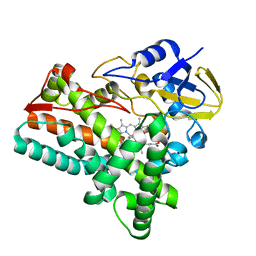 | | Crystal structure of cytochrome P450mel 107F1 in complex with heme and imidazole | | Descriptor: | Cytochrome P-450, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
5L4M
 
 | | Crystal Structure of Human Transthyretin in Complex with 3,5,6-Trichloro-2-pyridinyloxyacetic acid (Triclopyr) | | Descriptor: | SODIUM ION, Transthyretin, Triclopyr | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6KZT
 
 | | Crystal structure of cytochrome P450mel 107F1 with biaryl coupling reactivity | | Descriptor: | Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hou, X.D, Rao, Y.J, Icyishaka, P, Li, C.X, Lou, J.L. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray analysis of cytochrome P450mel 107F1 with biaryl coupling reactivity
To Be Published
|
|
6L1W
 
 | | Zinc-finger Antiviral Protein (ZAP) bound to RNA | | Descriptor: | RNA (5'-R(*CP*GP*UP*CP*GP*U)-3'), ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Luo, X, Wang, X, Gao, Y, Zhu, J, Liu, S, Gao, G, Gao, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein.
Cell Rep, 30, 2020
|
|
