6ULP
 
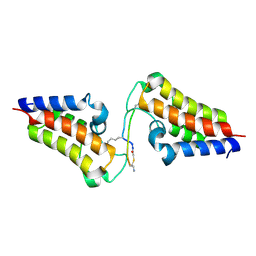 | | BRD3-BD2 in complex with the cyclic peptide 3.2_3 | | Descriptor: | Bromodomain-containing protein 3, Cyclic peptide 3.2_3 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-10-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7Z31
 
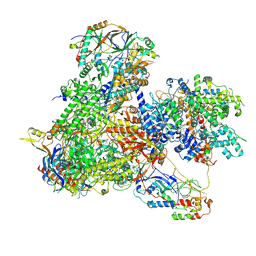 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7AQ8
 
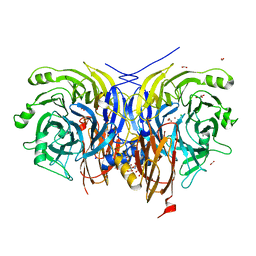 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y/D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1PFK
 
 | |
6XRI
 
 | |
7Z30
 
 | |
6OOG
 
 | |
2TUN
 
 | |
7Z9P
 
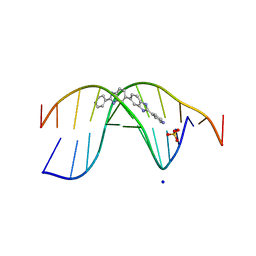 | | The novel DNA binding mechanism of ridinilazole, a precision Clostridiodes difficile antibiotic | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*(RID))-3'), Ridinilazole, SODIUM ION, ... | | Authors: | Mason, S, Leonard, P.M. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Novel DNA Binding Mechanism of Ridinilazole, a Precision Clostridiodes difficile Antibiotic.
Antimicrob.Agents Chemother., 67, 2023
|
|
7AQ5
 
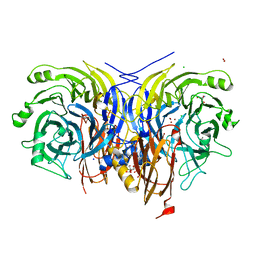 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583N | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7AQ6
 
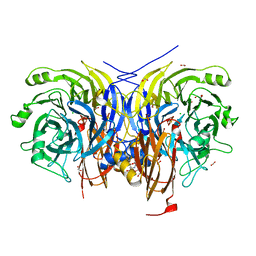 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583F | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
6OJA
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its L-methionine bound conformation | | Descriptor: | Lipoprotein, METHIONINE | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
6I3R
 
 | | Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1/2 of Drosophila helicase MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Buelow, S.v, Masiewicz, P, Simon, B, Hennig, J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res., 47, 2019
|
|
6OK2
 
 | |
6V7B
 
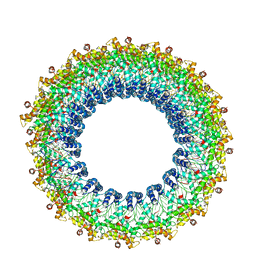 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
7AQ7
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
6OMT
 
 | | HIV-1 capsid hexamer R18D mutant | | Descriptor: | Capsid protein | | Authors: | Huang, P, Summers, B.J, Xiong, Y. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-21 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | FEZ1 Is Recruited to a Conserved Cofactor Site on Capsid to Promote HIV-1 Trafficking.
Cell Rep, 28, 2019
|
|
7AW6
 
 | | The extracellular region of CD33 with bound sialoside analogue P22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Thompson, A.P, Arrowsmith, C.H, Edwards, A.M, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The extracellular region of CD33 with bound sialoside analogue P22
To Be Published
|
|
6YA6
 
 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
9FMD
 
 | |
6AME
 
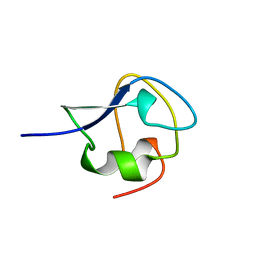 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1PCF
 
 | |
8CGW
 
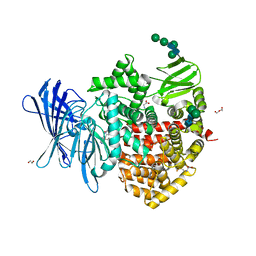 | | Insulin-regulated aminopeptidase (IRAP) in complex with an allosteric benzopyran-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanisms of Allosteric Inhibition of Insulin-Regulated Aminopeptidase.
J.Mol.Biol., 436, 2024
|
|
7AQ3
 
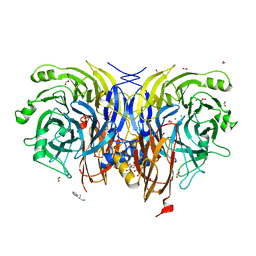 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583D | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
