4R4N
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-1 93ug037 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, Antibody 2.2c heavy CHAIN, ... | | Authors: | Acharya, P, Louder, R, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
130D
 
 | | MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG) COMPLEXED WITH HOECHST 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
6N4E
 
 | | hPGDS complexed with a quinoline-3-carboxamide | | Descriptor: | 7-(difluoromethoxy)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]quinoline-3-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
6W9E
 
 | | Crystal Structure of Human CDK9/cyclinT1 in complex with MC180295 | | Descriptor: | (4-amino-2-{[(1R,2R,4S)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, (4-amino-2-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, Cyclin-T1, ... | | Authors: | Zhang, P, Wu, J. | | Deposit date: | 2020-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Modeling of CDK9 Inhibitors to Explore Selectivity and Structure-Activity Relationships
To Be Published
|
|
6N81
 
 | |
7PET
 
 | | The 4x177 nucleosome array containing H1 | | Descriptor: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | Authors: | Dombrowski, M, Cramer, P. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4R6Q
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8CK6
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
13PK
 
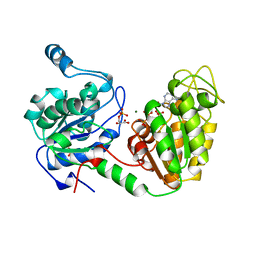 | | TERNARY COMPLEX OF PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI | | Descriptor: | 3-PHOSPHOGLYCERATE KINASE, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bernstein, B.E, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 1996-11-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation.
Nature, 385, 1997
|
|
8CKQ
 
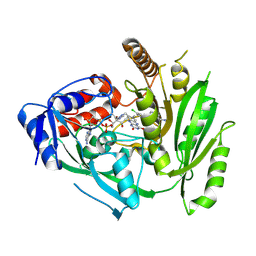 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
8CKT
 
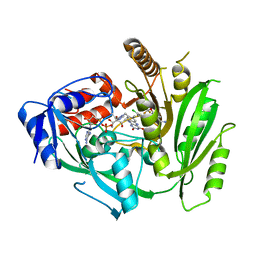 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
8CLW
 
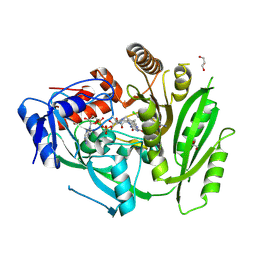 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
8CM2
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[[3,5-dichloro-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
7PX1
 
 | | Conotoxin from Conus mucronatus | | Descriptor: | CADMIUM ION, Conus mucronatus, IODIDE ION | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
4REB
 
 | | Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
7PX2
 
 | | Conotoxin Mu8.1 from Conus mucronatus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Conotoxin Mu8.1 | | Authors: | Mueller, E, Hackney, C, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
1A5C
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM PLASMODIUM FALCIPARUM | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Kim, H, Certa, U, Dobeli, H, Jakob, P, Hol, W.G.J. | | Deposit date: | 1998-02-13 | | Release date: | 1998-06-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum.
Biochemistry, 37, 1998
|
|
7Q4V
 
 | | Electron bifurcating hydrogenase - HydABC from A. woodii | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Katsyv, A, Kumar, A, Saura, P, Poeverlein, M.C, Freibert, S.A, Stripp, S, Jain, S, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
7Q4W
 
 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
1A1R
 
 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
5K77
 
 | | Dbr1 in complex with 7-mer branched RNA | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6WY3
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; P212121 form | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*(LV2)P*GP*CP*CP*GP*G)-3') | | Authors: | Sekula, B, Ruszkowski, M, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6Y0P
 
 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
