3SI7
 
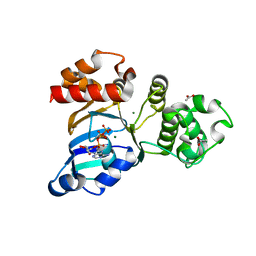 | | The crystal structure of the NBD1 domain of the mouse CFTR protein, deltaF508 mutant | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Brautigam, C.A, Caspa, E, Thomas, P.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Requirements for efficient correction of DeltaF508 CFTR revealed by analyses of evolved sequences
Cell(Cambridge,Mass.), 148, 2012
|
|
7FSB
 
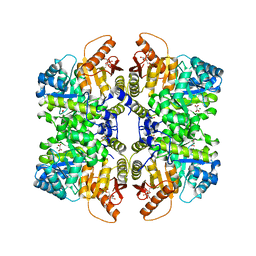 | | Structure of liver pyruvate kinase in complex with allosteric modulator 41 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRW
 
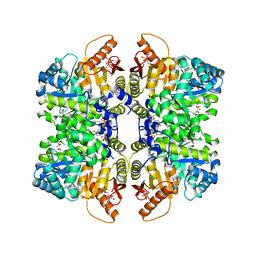 | | Structure of liver pyruvate kinase in complex with allosteric modulator 4 | | Descriptor: | (10aM)-3,8,9-trihydroxy-6H-6lambda~6~-dibenzo[c,e][1,2]oxathiine-6,6-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS4
 
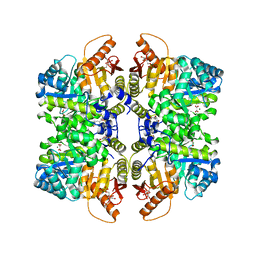 | | Structure of liver pyruvate kinase in complex with allosteric modulator 16 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4,4',5-trihydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}[1,1'-biphenyl]-2-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
4DGF
 
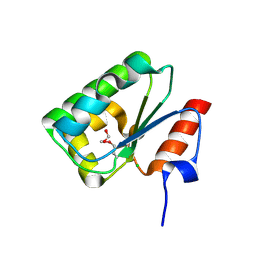 | | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution | | Descriptor: | CHLORIDE ION, FORMIC ACID, SULFATE TRANSPORTER SULFATE TRANSPORTER FAMILY PROTEIN | | Authors: | Keller, J.P, Chang, C, Tesar, C, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution
To be Published
|
|
7FS9
 
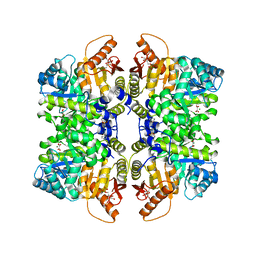 | | Structure of liver pyruvate kinase in complex with allosteric modulator 22 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(4-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSD
 
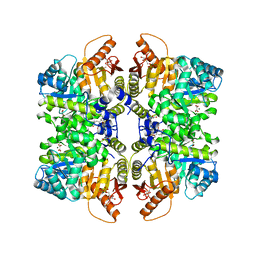 | | Structure of liver pyruvate kinase in complex with allosteric modulator 44 | | Descriptor: | (1M)-N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)-3',4,4',5-tetrahydroxy[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS2
 
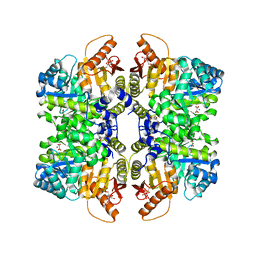 | | Structure of liver pyruvate kinase in complex with allosteric modulator 13 | | Descriptor: | (1P)-3',4,4',5-tetrahydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS8
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 21 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(3-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSC
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 42 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)benzenesulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
2WF7
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8IYH
 
 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
1XN0
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With (R,S)-Rolipram | | Descriptor: | MAGNESIUM ION, ROLIPRAM, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1Y2J
 
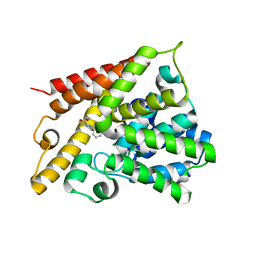 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With 3,5-dimethyl-1-(3-nitro-phenyl)-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 3,5-DIMETHYL-1-(3-NITROPHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
5C1M
 
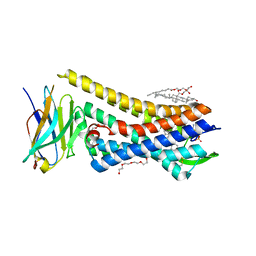 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
6XXQ
 
 | |
8JIX
 
 | |
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
1D6Y
 
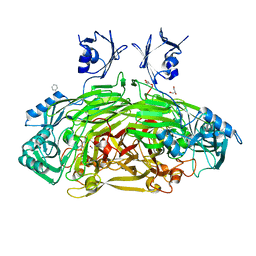 | | CRYSTAL STRUCTURE OF E. COLI COPPER-CONTAINING AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE AND COMPLEXED WITH NITRIC OXIDE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
2VTO
 
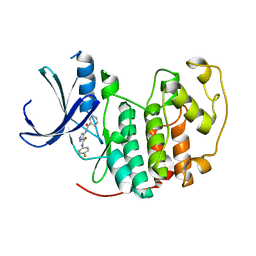 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(4-FLUOROPHENYL)-4-[(PHENYLCARBONYL)AMINO]-1H-PYRAZOLE-3-CARBOXAMIDE | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
1J9V
 
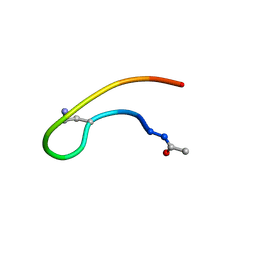 | | Solution structure of a lactam analogue (DabD) of HIV gp41 600-612 loop. | | Descriptor: | DabD (Ace)IWG(DAB)SGKLIDTTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-29 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1T46
 
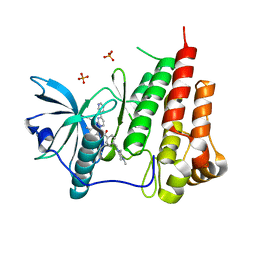 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
8IYW
 
 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
4DJI
 
 | | Structure of glutamate-GABA antiporter GadC | | Descriptor: | Probable glutamate/gamma-aminobutyrate antiporter | | Authors: | Ma, D, Lu, P.L, Yan, C.Y, Fan, C, Yin, P, Wang, J.W, Shi, Y.G. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure and mechanism of a glutamate-GABA antiporter
Nature, 483, 2012
|
|
3ZHQ
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MULTIFUNCTIONAL 2-OXOGLUTARATE METABOLISM ENZYME, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
