5O6V
 
 | |
6HCL
 
 | | Crystal structure of a MFS transporter with Ligand at 2.69 Angstroem resolution | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Major facilitator superfamily MFS_1, nonyl beta-D-glucopyranoside | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2018-08-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of L-lactate transport in the SLC16 solute carrier family.
Nat Commun, 10, 2019
|
|
8CHK
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
6WAO
 
 | |
8CHQ
 
 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
1U07
 
 | | Crystal Structure of the 92-residue C-term. part of TonB with significant structural changes compared to shorter fragments | | Descriptor: | TonB protein | | Authors: | Koedding, J, Killig, F, Polzer, P, Howard, S.P, Diederichs, K, Welte, W. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of a 92-residue c-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments
J.Biol.Chem., 280, 2005
|
|
3HZ2
 
 | |
3HZB
 
 | |
6HHT
 
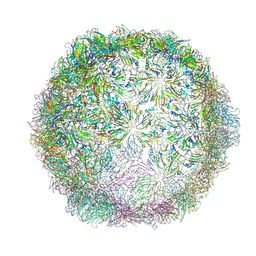 | | Echovirus 18 Open particle without two pentamers | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
5O9G
 
 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
6NWS
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1EPM
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS2, THR-PHE-GLN-ALA-PSA-LEU-ARG-GLU, ... | | Authors: | Crawford, M, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
8U1D
 
 | | Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1285 Heavy Chain, DH1285 Light Chain, ... | | Authors: | Thakur, B, Stalls, V.D, Acharya, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques.
Cell, 187, 2024
|
|
1EQC
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS IN COMPLEX WITH CASTANOSPERMINE AT 1.85 A | | Descriptor: | CASTANOSPERMINE, EXO-(B)-(1,3)-GLUCANASE | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
6VK2
 
 | |
4H9O
 
 | | Complex structure 2 of DAXX/H3.3(sub5,G90M)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
6BVE
 
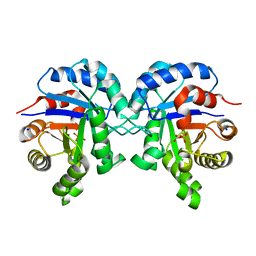 | |
6WCX
 
 | |
4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
5FWH
 
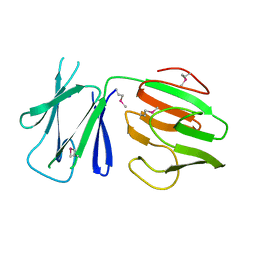 | | N-terminal FHA domain from EssC a component of the bacterial Type VII secretion apparatus | | Descriptor: | ESSC | | Authors: | Zoltner, M, Ng, W.M.A.V, Money, J.J, Fyfe, P.K, Kneuper, H, Palmer, T, Hunter, W.N. | | Deposit date: | 2016-02-17 | | Release date: | 2016-04-06 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Essc: Domain Structures Inform on the Elusive Translocation Channel in the Type Vii Secretion System.
Biochem.J., 473, 2016
|
|
3CSK
 
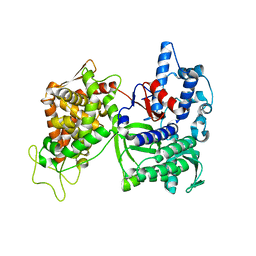 | | Structure of DPP III from Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, Probable dipeptidyl-peptidase 3, ZINC ION | | Authors: | Baral, P.K, Jajcanin, N, Deller, S, Macheroux, P, Abramic, M, Gruber, K. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding.
J.Biol.Chem., 283, 2008
|
|
6NBS
 
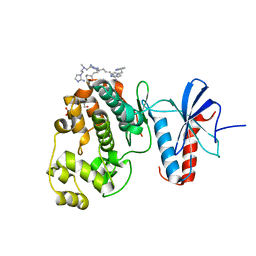 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
3MX0
 
 | | Crystal Structure of EphA2 ectodomain in complex with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 2, ... | | Authors: | Himanen, J.P, Yermekbayeva, L, Janes, P.W, Walker, J.R, Xu, K, Atapattu, L, Rajashankar, K.R, Mensinga, A, Lackmann, M, Nikolov, D.B, Dhe-Paganon, S. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3CY5
 
 | |
