8F45
 
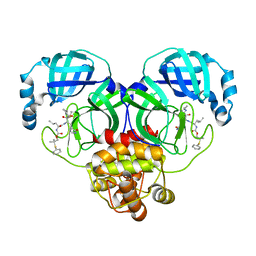 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl dimethyl sulfane inhibitor (cyclopropyl ketoamide warhead) | | Descriptor: | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
5L7P
 
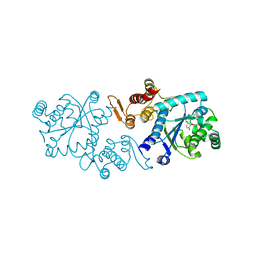 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
8F46
 
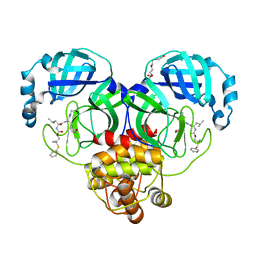 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor (cyano warhead) | | Descriptor: | 3C-like proteinase, N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
4O0O
 
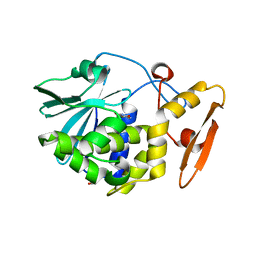 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution
To be Published
|
|
3MGN
 
 | | D-Peptide inhibitor PIE71 in complex with IQN17 | | Descriptor: | D-PEPTIDE INHIBITOR PIE71, IQN17 | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Francis, N. | | Deposit date: | 2010-04-07 | | Release date: | 2011-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
1YG4
 
 | |
2UWL
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8F44
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
5LAL
 
 | | Structure of Arabidopsis dirigent protein AtDIR6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dirigent protein 6, ... | | Authors: | Gasper, R, Kolesinski, P, Terlecka, B, Effenberger, I, Schaller, A, Hofmann, E. | | Deposit date: | 2016-06-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6.
Plant Physiol., 172, 2016
|
|
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3MME
 
 | | Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 HEAVY CHAIN FAB, ... | | Authors: | Pancera, M, McLellan, J, Zhou, T, Zhu, J, Kwong, P. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
1UM6
 
 | | catalytic antibody 21h3 | | Descriptor: | antibody 21h3, H chain, L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
2V5X
 
 | | Crystal structure of HDAC8-inhibitor complex | | Descriptor: | (2R)-N~8~-HYDROXY-2-{[(5-METHOXY-2-METHYL-1H-INDOL-3-YL)ACETYL]AMINO}-N~1~-[2-(2-PHENYL-1H-INDOL-3-YL)ETHYL]OCTANEDIAMIDE, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C, Gallinari, P, Jones, P, Mattu, M, Carfi, A, Defrancesco, R, Steinkuhler, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
1KRI
 
 | |
3V13
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
1V07
 
 | | CRYSTAL STRUCTURE OF ThrE11Val mutant of THE NERVE TISSUE MINI-HEMOGLOBIN FROM THE NEMERTEAN WORM CEREBRATULUS LACTEUS | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Ascenzi, P, Geuens, E, Dewilde, S, Moens, L, Bolognesi, M, Riggs, A, Hale, A, Deng, P, Nienhaus, G.U, Olson, J.S, Nienhaus, K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thr-E11 Regulates O2 Affinity in Cerebratulus Lacteus Mini-Hemoglobin
J.Biol.Chem., 279, 2004
|
|
6EIH
 
 | |
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
4C0U
 
 | | Cryo-EM reconstruction of enterovirus 71 in complex with a neutralizing antibody E18 | | Descriptor: | FAB EV18 4 D6-1 F1 G9, VP1, VP2, ... | | Authors: | Plevka, P, Perera, R, Cardosa, J, Suksatu, A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2013-08-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5OPK
 
 | | Crystal structure of D52N/R367Q cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
4G19
 
 | | Crystal structure of the glutathione transferase GTE1 from Phanerochaete chrysosporium in complex with glutathione | | Descriptor: | ACETATE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Didierjean, C, Favier, F, Prosper, P. | | Deposit date: | 2012-07-10 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Phanerochaete chrysosporium Glutathione Transferase Reveals a Novel Structural and Functional Class with Ligandin Properties.
J.Biol.Chem., 287, 2012
|
|
1URM
 
 | | HUMAN PEROXIREDOXIN 5, C47S MUTANT | | Descriptor: | BENZOIC ACID, CHLORIDE ION, PEROXIREDOXIN 5 | | Authors: | Declercq, J.P, Evrard, C. | | Deposit date: | 2003-10-31 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the C47S Mutant of Human Peroxiredoxin 5
J. Chem. Cryst., 34, 2004
|
|
1YFH
 
 | | wt Human O6-Alkylguanine-DNA Alkyltransferase Bound To DNA Containing an Alkylated Cytosine | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*AP*CP*AP*CP*AP*TP*CP*CP*AP*CP*A)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*(XCY)P*GP*TP*GP*TP*AP*GP*GP*T)-3', Methylated-DNA--protein-cysteine methyltransferase, ... | | Authors: | Duguid, E.M, Rice, P.A, He, C. | | Deposit date: | 2004-12-31 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The structure of the human AGT protein bound to DNA and its implications for damage detection.
J.Mol.Biol., 350, 2005
|
|
2Q5G
 
 | | Ligand binding domain of PPAR delta receptor in complex with a partial agonist | | Descriptor: | Peroxisome proliferator-activated receptor delta, [(7-{[2-(3-MORPHOLIN-4-YLPROP-1-YN-1-YL)-6-{[4-(TRIFLUOROMETHYL)PHENYL]ETHYNYL}PYRIDIN-4-YL]THIO}-2,3-DIHYDRO-1H-INDEN- 4-YL)OXY]ACETIC ACID | | Authors: | Pettersson, I, Sauerberg, P, Johansson, E, Hoffman, I, Tari, L.W, Hunter, M.J, Nix, J. | | Deposit date: | 2007-06-01 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of a partial PPARdelta agonist.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3V04
 
 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | 4-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-1H-indazole-5-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
