7MDS
 
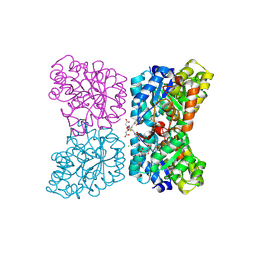 | | Crystal structure of AtDHDPS1 in complex with MBDTA-2 | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase 1, chloroplastic, CHLORIDE ION, ... | | Authors: | Hall, C.J, Soares da Costa, T.P, Panjikar, S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants.
Elife, 10, 2021
|
|
7MPG
 
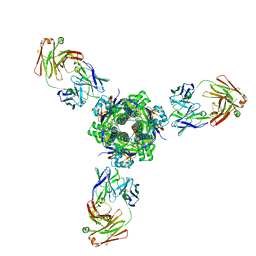 | |
8BXU
 
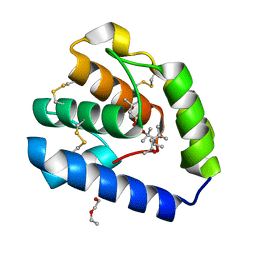 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXV
 
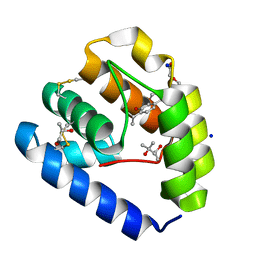 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Thymol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-METHYL-2-(1-METHYLETHYL)PHENOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXW
 
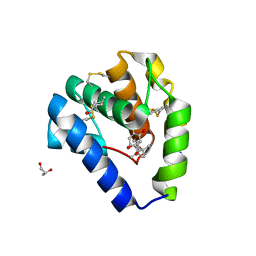 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Carvacrol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-methyl-5-propan-2-yl-phenol, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
1AGB
 
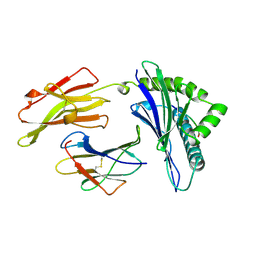 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
7MH2
 
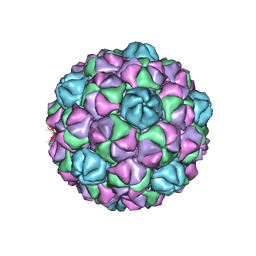 | |
7MJ1
 
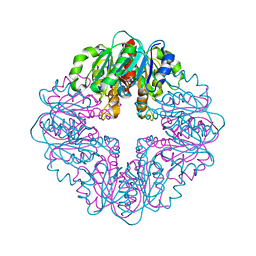 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with NAD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1A7T
 
 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
1AMM
 
 | | 1.2 ANGSTROM STRUCTURE OF GAMMA-B CRYSTALLIN AT 150K | | Descriptor: | GAMMA B-CRYSTALLIN | | Authors: | Kumaraswamy, V.S, Lindley, P.F, Slingsby, C, Glover, I.D. | | Deposit date: | 1996-03-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An eye lens protein-water structure: 1.2 A resolution structure of gammaB-crystallin at 150 K.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1ACX
 
 | |
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BAP
 
 | |
1AVN
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH THE HISTAMINE ACTIVATOR | | Descriptor: | AZIDE ION, CARBONIC ANHYDRASE II, HISTAMINE, ... | | Authors: | Briganti, F, Mangani, S, Orioli, P, Scozzafava, A, Vernaglione, G, Supuran, C.T. | | Deposit date: | 1997-09-17 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine.
Biochemistry, 36, 1997
|
|
1B96
 
 | |
1B39
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 PHOSPHORYLATED ON THR 160 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (CELL DIVISION PROTEIN KINASE 2) | | Authors: | Brown, N.R, Noble, M.E.M, Lawrie, A.M, Morris, M.C, Tunnah, P, Divita, G, Johnson, L.N, Endicott, J.A. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity.
J.Biol.Chem., 274, 1999
|
|
1B97
 
 | |
1B20
 
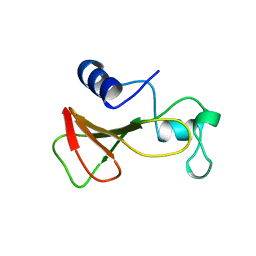 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3FPG
 
 | | Crystal Structure of E81Q mutant of MtNAS | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, Putative uncharacterized protein | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2009-01-05 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6TT2
 
 | |
3F29
 
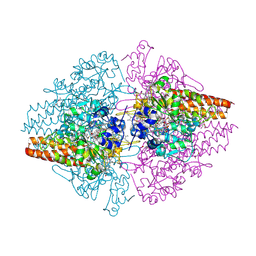 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-10-29 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
4N18
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
4N4U
 
 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3FNT
 
 | | Crystal structure of pepstatin A bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, HAP protein, Inhibitor, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
3F7K
 
 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
