4F9U
 
 | | Structure of glycosylated glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CG32412, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
7RH8
 
 | | Crystal structure of Fur1p from Candida albicans, in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uracil phosphoribosyltransferase | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Fur1p from Candida albicans, in complex with UTP
To Be Published
|
|
4TUH
 
 | | Bcl-xL in complex with inhibitor (Compound 10) | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydroisoquinolin-2(1H)-yl]-5-{3-[4-(1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenoxy]propyl}-1,3-thiazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Czabotar, P.E, Lessense, G, Smith, B.J, Colman, P.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Rescaffolding of Selective Antagonists of BCL-XL.
Acs Med.Chem.Lett., 5, 2014
|
|
4FAK
 
 | | Crystal Structure of OrfX in Complex with S-Adenosylmethionine | | Descriptor: | PHOSPHATE ION, Ribosomal RNA large subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | Authors: | Safo, M.K, Musayev, F.N, Boundy, S, Archer, G.L, Rife, J.P, O'Farrell, H.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the Staphylococcus aureus rRNA Methyltransferase Encoded by orfX, the Gene Containing the Staphylococcal Chromosome Cassette mec (SCCmec) Insertion Site.
J.Biol.Chem., 288, 2013
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
3UO5
 
 | | Aurora A in complex with YL1-038-31 | | Descriptor: | 4-{[4-(phenylamino)pyrimidin-2-yl]amino}benzoic acid, Serine/Threonine-Protein Kinase 6 | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7012 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
3UOD
 
 | | Aurora A in complex with RPM1693 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[2-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzoic acid, Aurora kinase A, ... | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5002 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
7RLL
 
 | | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate | | Descriptor: | Arf3p, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stogios, P.J, Michalska, K, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-25 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate
To Be Published
|
|
5KH9
 
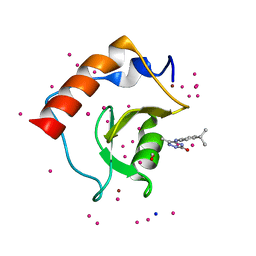 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5KHP
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
1LZH
 
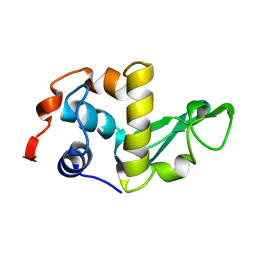 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3HJH
 
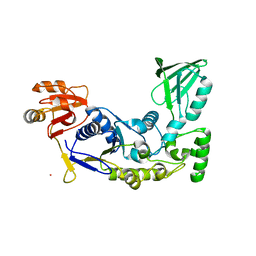 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
7RT0
 
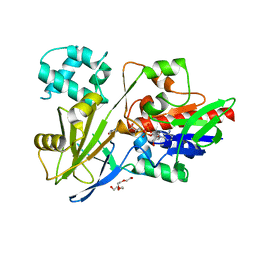 | |
4FLO
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
3HG1
 
 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
5KHR
 
 | | Model of human Anaphase-promoting complex/Cyclosome complex (APC15 deletion mutant) in complex with the E2 UBE2C/UBCH10 poised for ubiquitin ligation to substrate (APC/C-CDC20-substrate-UBE2C) | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | VanderLinden, R, Yamaguchi, M, Dube, P, Haselbach, D, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
1M0F
 
 | | Structural Studies of Bacteriophage alpha3 Assembly, Cryo-electron microscopy | | Descriptor: | Capsid Protein F, Major Spike Protein G, Scaffolding Protein B, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V.D, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
4FOL
 
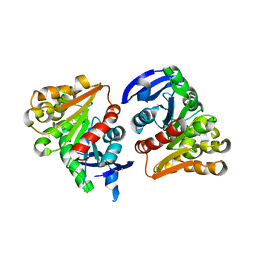 | | S-formylglutathione hydrolase Variant H160I | | Descriptor: | S-formylglutathione hydrolase | | Authors: | Legler, P.M, Millard, C.B. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A role for His-160 in peroxide inhibition of S. cerevisiae S-formylglutathione hydrolase: Evidence for an oxidation sensitive motif.
Arch.Biochem.Biophys., 528, 2012
|
|
3UVN
 
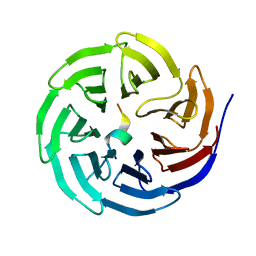 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
4FP7
 
 | |
4FOW
 
 | |
1MRY
 
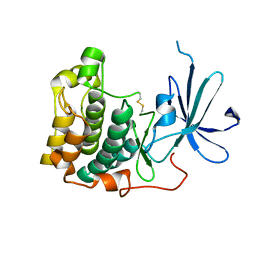 | | crystal structure of an inactive akt2 kinase domain | | Descriptor: | RAC-beta serine/threonine kinase | | Authors: | Huang, X, Begley, M, Morgenstern, K.A, Gu, Y, Rose, P, Zhao, H, Zhu, X. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an inactive akt2 kinase domain
Structure, 11, 2003
|
|
3V1Y
 
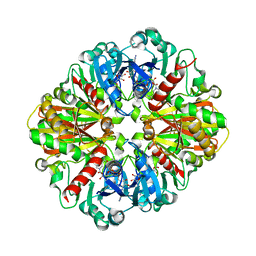 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
6KNA
 
 | |
1MS1
 
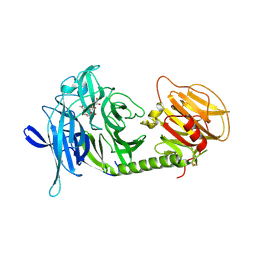 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
