7TEI
 
 | |
6VCF
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, iron form | | Descriptor: | BICARBONATE ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7TF8
 
 | |
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6FWZ
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-07 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5A85
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 8-(3R,4R)-3-(cyclohexylmethoxy)piperidin-4-ylamino-3-methyl-1,2-dihydro-1,7- naphthyridin-2-one | | Descriptor: | (3R,4R)-3-(cyclohexylmethoxy)piperidin-4-yl]amino}-3-methyl-1,2-dihydro-1,7-naphthyridin-2-one, 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Based Optimization of Naphthyridones Into Potent Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5SYB
 
 | | Crystal structure of human PHF5A | | Descriptor: | 1,2-ETHANEDIOL, PHD finger-like domain-containing protein 5A, ZINC ION | | Authors: | Tsai, J.H.C, Teng, T, Zhu, P, Fekkes, P, Larsen, N.A. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex.
Nat Commun, 8, 2017
|
|
1FZK
 
 | | MHC CLASS I NATURAL MUTANT H-2KBM1 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
5AJO
 
 | | Crystal structure of the inactive form of GalNAc-T2 in complex with the glycopeptide MUC5AC-3,13 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUCIN, POLYPEPTIDE N-ACETYLGALACTOSAMINYLTRANSFERASE 2, ... | | Authors: | Lira-Navarrete, E, delasRivas, M, Companon, I, Pallares, M.C, Kong, Y, Iglesias-Fernandez, J, Bernardes, G.J.L, Peregrina, J.M, Rovira, C, Bernado, P, Bruscolini, P, Clausen, H, Lostao, A, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Nat.Commun., 6, 2015
|
|
7A1Z
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.024 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6MUV
 
 | | The structure of the Plasmodium falciparum 20S proteasome in complex with two PA28 activators | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
5GRO
 
 | | Crystal structure of the N-terminal anticodon-binding domain of non-discriminating aspartyl-tRNA synthetase from Helicobacter pylori | | Descriptor: | Aspartate--tRNA(Asp/Asn) ligase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Songsiriritthigul, C, Fuengfuloy, P, Chen, C.-J, Suebka, S, Chuawong, P. | | Deposit date: | 2016-08-12 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N-terminal anticodon-binding domain of the nondiscriminating aspartyl-tRNA synthetase from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7A6C
 
 | |
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
6OHK
 
 | |
6ZNL
 
 | | Cryo-EM structure of the dynactin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
6ZMM
 
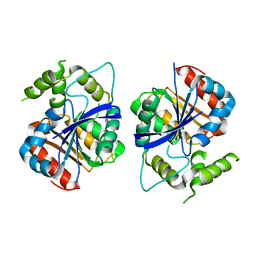 | | Crystal structure of human NDRG1 | | Descriptor: | CHLORIDE ION, Protein NDRG1 | | Authors: | Mustonen, V, Kursula, P, Ruskamo, S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal and solution structure of NDRG1, a membrane-binding protein linked to myelination and tumour suppression.
Febs J., 288, 2021
|
|
5JVF
 
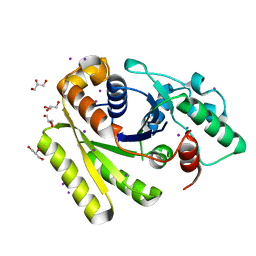 | | Crystal Structure of Apo-FleN | | Descriptor: | GLYCEROL, IODIDE ION, Site-determining protein | | Authors: | Jain, D, Chanchal, Banerjee, P. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ATP-Induced Structural Remodeling in the Antiactivator FleN Enables Formation of the Functional Dimeric Form
Structure, 25, 2017
|
|
8DBA
 
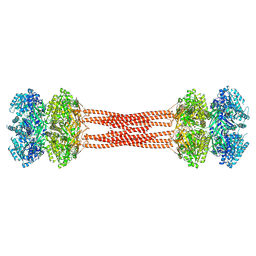 | | Crystal structure of dodecameric KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
1G9M
 
 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
6VGC
 
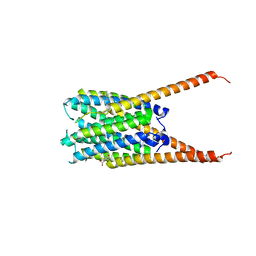 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
5YGU
 
 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
7XBY
 
 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
1G86
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN/N-ETHYLMALEIMIDE COMPLEX | | Descriptor: | CHARCOT-LEYDEN CRYSTAL PROTEIN, N-ETHYLMALEIMIDE | | Authors: | Ackerman, S.J, Liu, L, Kwatia, M.A, Savage, M.P, Leonidas, D.D, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion.
J.Biol.Chem., 277, 2002
|
|
4BOS
 
 | | Structure of OTUD2 OTU domain in complex with Ubiquitin K11-linked peptide | | Descriptor: | MAGNESIUM ION, NITRATE ION, OTUD2, ... | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
