4XBD
 
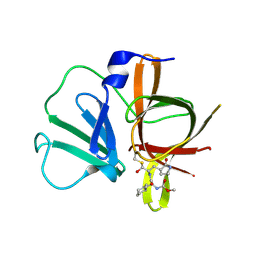 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
8PIM
 
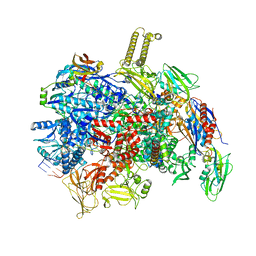 | | fully recruited RfaH bound to E. coli transcription complex paused at ops site (not complementary scaffold) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
7PUG
 
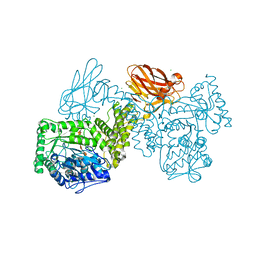 | | GH115 alpha-1,2-glucuronidase in complex with xylopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-09-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8TXY
 
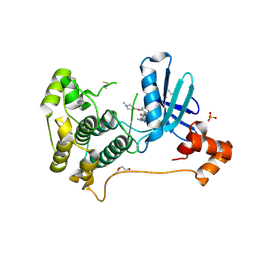 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6YPZ
 
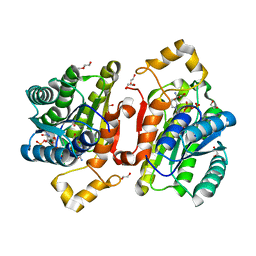 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
8PIB
 
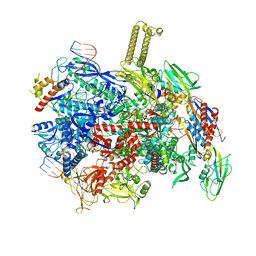 | | autoinhibited RfaH bound to E. coli transcription complex paused at ops site (encounter complex) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-21 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PFG
 
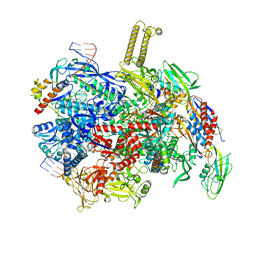 | | autoinhibited RfaH bound to E. coli transcription complex paused at ops site (encounter complex), not fully complementary scaffold | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
5F4N
 
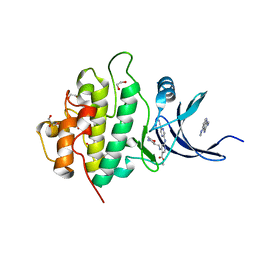 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | Authors: | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
6P6K
 
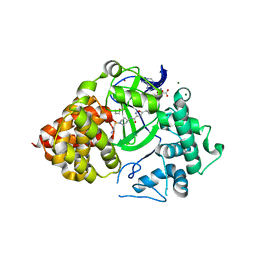 | |
4XOJ
 
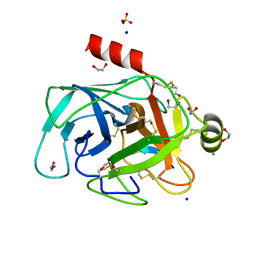 | | Structure of bovine trypsin in complex with analogues of sunflower inhibitor 1 (SFTI-1) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Golik, P, Malicki, S, Grudnik, P, Karna, N, Debowski, D, Legowska, A, Wladyka, B, Gitlin, A, Brzozowski, K, Dubin, G, Rolka, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Investigation of Serine-Proteinase-Catalyzed Peptide Splicing in Analogues of Sunflower Trypsin Inhibitor 1 (SFTI-1).
Chembiochem, 16, 2015
|
|
8PHK
 
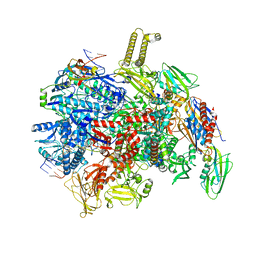 | | fully recruited RfaH bound to E. coli transcription complex paused at ops site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
4XOU
 
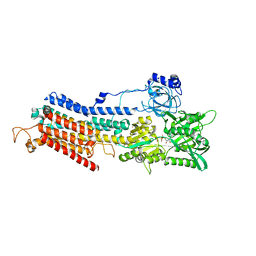 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
6YAQ
 
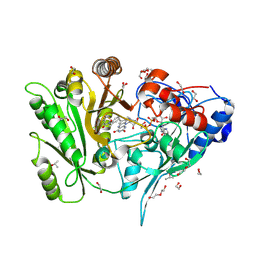 | | Crystal sttructure of ZmCKO8 in complex with inhibitor 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea, Cytokinin dehydrogenase 8, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
5MDJ
 
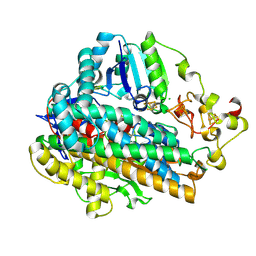 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in a its as-isolated high-pressurized form | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6P0U
 
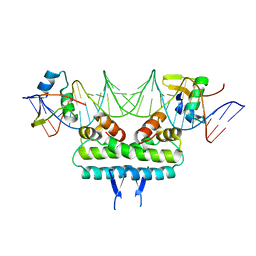 | | Crystal structure of ternary DNA complex " FX(1-2)-2Xis" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
7AL1
 
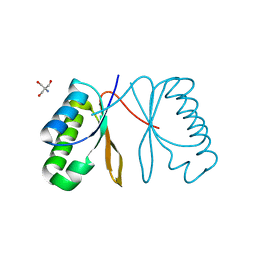 | | Cell division protein SepF from Methanobrevibacter smithii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein SepF | | Authors: | Sogues, A, wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SepF is the FtsZ anchor in archaea, with features of an ancestral cell division system.
Nat Commun, 12, 2021
|
|
7AHM
 
 | | Low-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, K(+)/H(+) antiporter subunit KhtT | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PE1
 
 | |
7PE2
 
 | |
5VP2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with madumycin II and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Osterman, I.A, Khabibullina, N.F, Komarova, E.S, Kasatsky, P, Kartsev, V.G, Bogdanov, A.A, Dontsova, O.A, Konevega, A.L, Sergiev, P.V, Polikanov, Y.S. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state.
Nucleic Acids Res., 45, 2017
|
|
7PR3
 
 | | Cocrystal Form I of a cytochrome c, sulfonato-thiacalix[4]arene - zinc cluster | | Descriptor: | Cytochrome c iso-1, HEME C, PHOSPHATE ION, ... | | Authors: | Flood, R.J, Ramberg, K, Guagnini, F, Crowley, P.B. | | Deposit date: | 2021-09-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Protein Frameworks with Thiacalixarene and Zinc.
Cryst.Growth Des., 22, 2022
|
|
7ZVX
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7ZVN
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 A resolution | | Descriptor: | 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, CALCIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
1AW5
 
 | |
7AOT
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
