7A7B
 
 | | Bacillithiol Disulfide Reductase Bdr (YpdA) from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YpdA family putative bacillithiol disulfide reductase Bdr | | Authors: | Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2020-08-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase.
Biochemistry, 59, 2020
|
|
7R49
 
 | | Crystal structure of the L. plantarum acyl carrier protein synthase (AcpS)in complex with D-alanyl carrier protein (DltC1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4'-PHOSPHOPANTETHEINE, D-alanyl carrier protein 1, ... | | Authors: | Nikolopoulos, N, Ravaud, S, Simorre, J.P, Grangeasse, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | DltC acts as an interaction hub for AcpS, DltA and DltB in the teichoic acid D-alanylation pathway of Lactiplantibacillus plantarum.
Sci Rep, 12, 2022
|
|
1AM4
 
 | | COMPLEX BETWEEN CDC42HS.GMPPNP AND P50 RHOGAP (H. SAPIENS) | | Descriptor: | CDC42HS, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 1997-06-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP.
Nature, 388, 1997
|
|
7MDO
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
5KMM
 
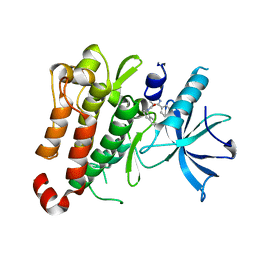 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(1-naphthyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-naphthalen-1-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8OM2
 
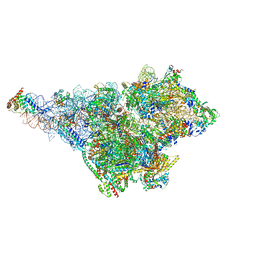 | | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
1AJR
 
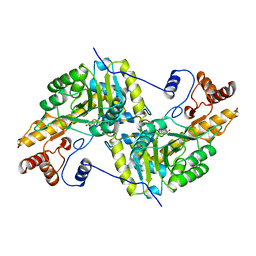 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
7RT7
 
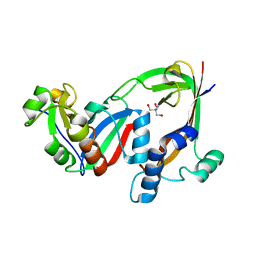 | | Crystal structure of the RhsP2 C-terminal toxin domain in complex with its immunity protein, RhsI2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhsI2, RhsP2 | | Authors: | Bullen, N.P, Prehna, G, Whitney, J.C. | | Deposit date: | 2021-08-12 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An ADP-ribosyltransferase toxin kills bacterial cells by modifying structured non-coding RNAs.
Mol.Cell, 82, 2022
|
|
8OM4
 
 | | Small subunit of yeast mitochondrial ribosome. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8BVI
 
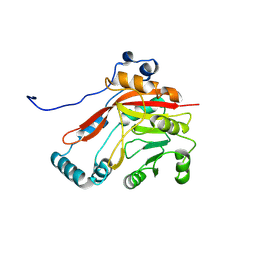 | |
6L6Y
 
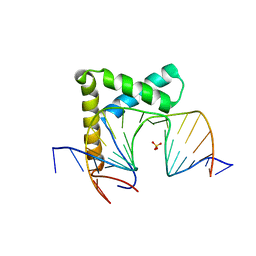 | |
6UAR
 
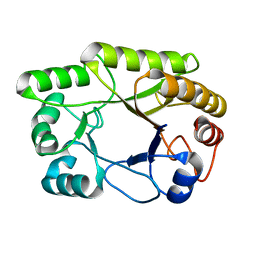 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAU
 
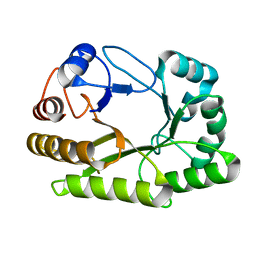 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glyco_hydro_cc domain-containing protein, ZINC ION, ... | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB0
 
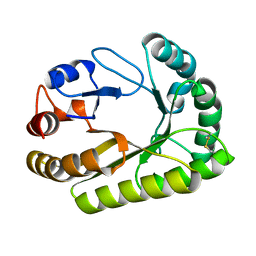 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
3HOW
 
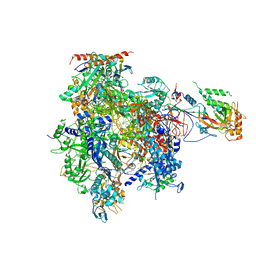 | | Complete RNA polymerase II elongation complex III with a T-U mismatch and a frayed RNA 3'-uridine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
5KMJ
 
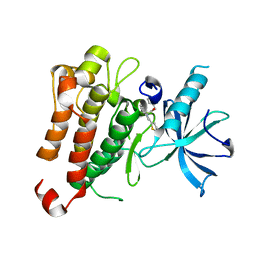 | | TrkA JM-kinase with {N}-(2-pyridylmethyl)-2-[2-(2-thienyl)indol-1-yl]acetamide | | Descriptor: | High affinity nerve growth factor receptor, ~{N}-(pyridin-2-ylmethyl)-2-(2-thiophen-2-ylindol-1-yl)ethanamide | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5HL0
 
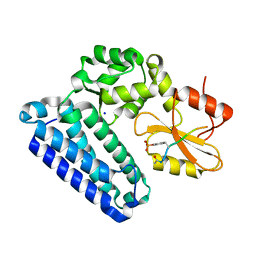 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
6UBB
 
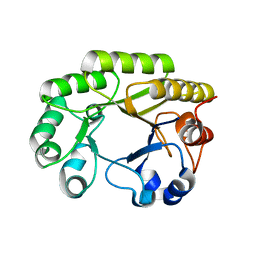 | | Crystal structure of a GH128 (subgroup VI) exo-beta-1,3-glucanase from Aureobasidium namibiae (AnGH128_VI) with laminaribiose at the surface-binding site | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Vieira, P.S, Domingues, M.N, Cordeiro, R.L, Tomazini, A, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6RAW
 
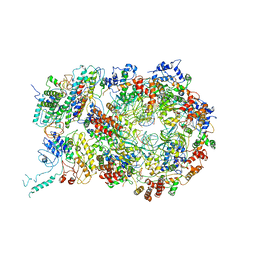 | | D. melanogaster CMG-DNA, State 1A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2019-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
7Z43
 
 | |
7A60
 
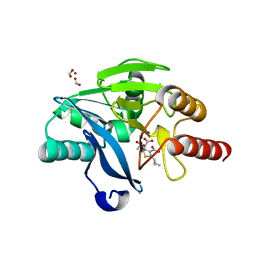 | | Crystal structure of VIM-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7Z42
 
 | |
7A61
 
 | | Crystal structure of KPC-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (2~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-butyl-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
