8USM
 
 | | FmlH Lectin Domain UTI89 - AM4085 | | Descriptor: | 4'-fluoro-6-(trifluoromethyl)[1,1'-biphenyl]-2-yl 2-acetamido-2-deoxy-beta-D-galactopyranoside, Fimbrial protein (Fragment) | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Maddirala, A.R, Janetka, J.W, Hultgren, S.J. | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Orally Bioavailable FmlH Lectin Antagonists as Treatment for Urinary Tract Infections.
J.Med.Chem., 67, 2024
|
|
1E2A
 
 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
6PQ2
 
 | |
1DVC
 
 | | SOLUTION NMR STRUCTURE OF HUMAN STEFIN A AT PH 5.5 AND 308K, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STEFIN A | | Authors: | Martin, J.R, Craven, C.J, Jerala, R, Kroon-Zitko, L, Zerovnik, E, Turk, V, Waltho, J.P. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of human stefin A.
J.Mol.Biol., 246, 1995
|
|
6OS0
 
 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OLK
 
 | |
6OQH
 
 | |
5A6R
 
 | | Crystal structure of the BTB domain of human KCTD17 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD17 | | Authors: | Pinkas, D.M, Sorrell, F, Sanvitale, C.E, Goubin, S, Williams, E, Newman, J.A, Pearce, N.M, Neshich, I, Pike, A.C.W, MacKenzie, A, Quigley, A, Faust, B, Carpenter, E.P, Tallant, C, Kopec, J, Chalk, R, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
6OMA
 
 | |
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LWN
 
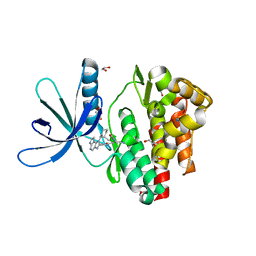 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | Descriptor: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
8EMA
 
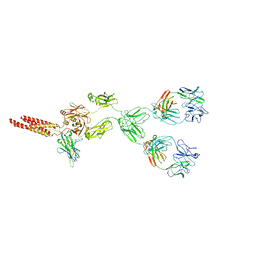 | | mouse full length B cell receptor | | Descriptor: | Anti-human Langerin 2G3 lambda chain, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Ying, D, Xiong, P, Michael, R. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
6PHG
 
 | |
2V3O
 
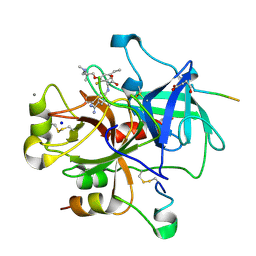 | | Thrombin with 3-cycle with F | | Descriptor: | (2R)-[(4-CARBAMIMIDOYLPHENYL)AMINO]{3-[3-(DIMETHYLAMINO)-2,2-DIMETHYLPROPOXY]-5-ETHYL-2-FLUOROPHENYL}ETHANOIC ACID, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Banner, D.W, Wessel, H.P. | | Deposit date: | 2007-06-19 | | Release date: | 2008-06-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fluorine in Pharmaceuticals: Looking Beyond Intuition.
Science, 317, 2007
|
|
7PZT
 
 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
6PDR
 
 | |
6PGM
 
 | | PirF geranyltransferase | | Descriptor: | PirF Geranyltransferase | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in Ribosomally Synthesized and Post-Translationally Modified Peptide Prenyltransferases.
J.Am.Chem.Soc., 140, 2018
|
|
6PIP
 
 | |
6PPC
 
 | | Solution structure of conotoxin MiXXVIIA | | Descriptor: | Conopeptide phi-MiXXVIIA | | Authors: | Daly, N.L, Dekan, Z, Jin, A.H, Alewood, P.F. | | Deposit date: | 2019-07-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conotoxin phi-MiXXVIIA from the Superfamily G2 Employs a Novel Cysteine Framework that Mimics Granulin and Displays Anti-Apoptotic Activity.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LWM
 
 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
5LYI
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(1~{R},2~{S},4~{R})-1,7,7-trimethyl-2-bicyclo[2.2.1]heptanyl]amino]propan-2-yl]sulfamoylamino]hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
6PQM
 
 | |
6PRQ
 
 | |
8OMG
 
 | | Crystal structure of mKHK (apo) | | Descriptor: | Ketohexokinase | | Authors: | Ebenhoch, E, Pautsch, P. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6PRJ
 
 | |
