7PMN
 
 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation II) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha-binding protein, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7PLO
 
 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7PFO
 
 | | Core human replisome | | Descriptor: | Cell division control protein 45 homolog, Claspin, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Jones, M.J, Yeeles, J.T.P. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human replisome shows the organisation and interactions of a DNA replication machine.
Embo J., 40, 2021
|
|
7PQD
 
 | | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 angstrom : the structural basis for dimerisation.
Biochem.J., 478, 2021
|
|
8TTF
 
 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
1CFB
 
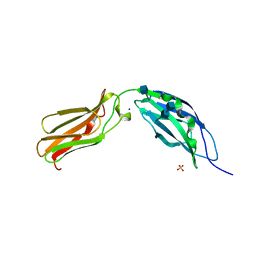 | | CRYSTAL STRUCTURE OF TANDEM TYPE III FIBRONECTIN DOMAINS FROM DROSOPHILA NEUROGLIAN AT 2.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DROSOPHILA NEUROGLIAN, ... | | Authors: | Huber, A.H, Wang, Y.E, Bieber, A.J, Bjorkman, P.J. | | Deposit date: | 1994-08-27 | | Release date: | 1994-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A.
Neuron, 12, 1994
|
|
1CPJ
 
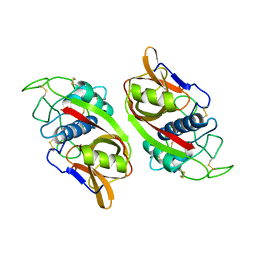 | |
8TTH
 
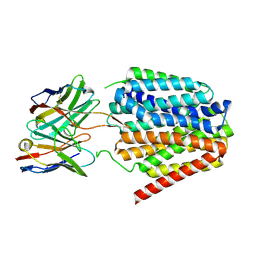 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTE
 
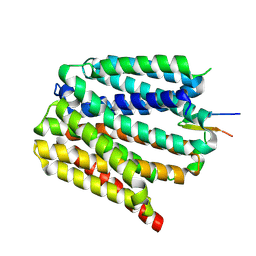 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTG
 
 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8K4S
 
 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-07-20 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch.
Cell, 2024
|
|
5ME8
 
 | | N-terminal domain of the human tumor suppressor ING5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Inhibitor of growth protein 5 | | Authors: | Roversi, P, Blanco, F.J, Rojas, A.L, Buitrago, J.A.R. | | Deposit date: | 2016-11-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
6OLJ
 
 | |
5MMT
 
 | |
6P2T
 
 | | BshB from Bacillus subtilis complexed with citrate | | Descriptor: | CITRIC ACID, N-acetyl-alpha-D-glucosaminyl L-malate deacetylase 1, SODIUM ION, ... | | Authors: | Cook, P.D, Meloche, C.E. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | X-ray crystallographic structure of BshB, the zinc-dependent deacetylase involved in bacillithiol biosynthesis.
Protein Sci., 29, 2020
|
|
6P3K
 
 | | Crystal structure of LigU(C100S) | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6PEL
 
 | |
1CTE
 
 | |
7PKR
 
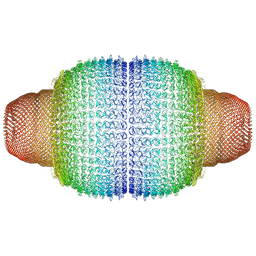 | | Vault structure in primmed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
1DB5
 
 | | HUMAN S-PLA2 IN COMPLEX WITH INDOLE 6 | | Descriptor: | 4-(1-BENZYL-3-CARBAMOYLMETHYL-2-METHYL-1H-INDOL-5-YLOXY)-BUTYRIC ACID, CALCIUM ION, PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Chirgadze, N.Y, Schevitz, R.W, Wery, J.-P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of the first potent and selective inhibitor of human non-pancreatic secretory phospholipase A2.
Nat.Struct.Biol., 2, 1995
|
|
8TK2
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with 2-((2,4-difluorobenzyl)amino)-2-oxoacetic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Dual specificity protein phosphatase 3, ... | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) binding 2-((2,4-difluorobenzyl)amino)-2-oxoacetic acid
To Be Published
|
|
7PKY
 
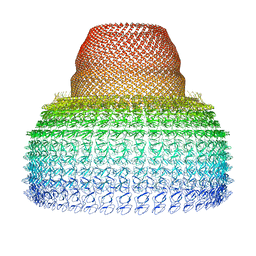 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
1DFY
 
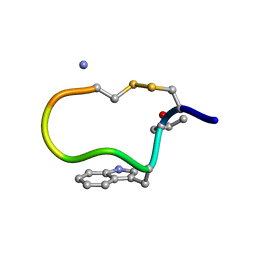 | | NMR STRUCTURE OF CONTRYPHAN-SM CYCLIC PEPTIDE (MAJOR FORM-CIS) | | Descriptor: | CONTRYPHAN-SM | | Authors: | Pallaghy, P.K, He, W, Jimenez, E.C, Olivera, B.M, Norton, R.S. | | Deposit date: | 1999-11-22 | | Release date: | 2002-05-01 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structures of the contryphan family of cyclic peptides. Role of electrostatic interactions in cis-trans isomerism.
Biochemistry, 39, 2000
|
|
1DAV
 
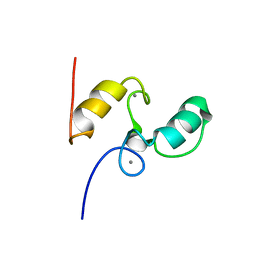 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DEI
 
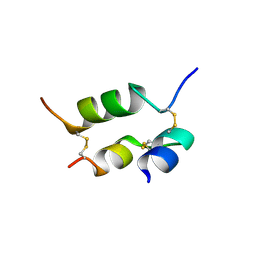 | | DESHEPTAPEPTIDE (B24-B30) INSULIN | | Descriptor: | INSULIN | | Authors: | Bao, S.-J, Chang, W.-R, Wan, Z.-L, Zhang, J.-P, Liang, D.-C. | | Deposit date: | 1996-05-15 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of desheptapeptide(B24-B30)insulin at 1.6 A resolution: implications for receptor binding.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
