3I0M
 
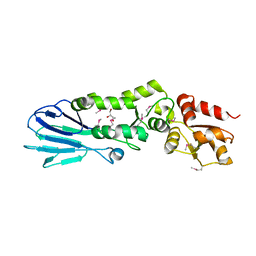 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
1KR8
 
 | |
4DAE
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 6-chloroguanosine | | Descriptor: | 6-chloro-9-(beta-D-ribofuranosyl)-9H-purin-2-amine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAN
 
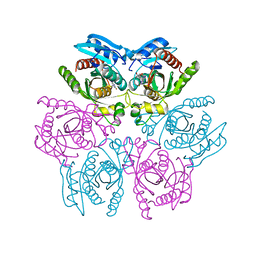 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4I85
 
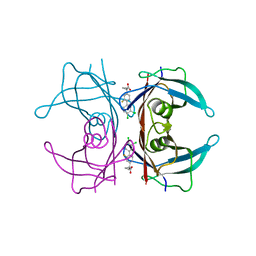 | | Crystal structure of transthyretin in complex with CHF5074 at neutral pH | | Descriptor: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, Transthyretin | | Authors: | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
3DWF
 
 | | Crystal Structure of the Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 Mutant F278E | | Descriptor: | 11-beta-hydroxysteroid dehydrogenase 1, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lawson, A.J, Ride, J.P, White, S.A, Walker, E.A. | | Deposit date: | 2008-07-22 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutations of key hydrophobic surface residues of 11 beta-hydroxysteroid dehydrogenase type 1 increase solubility and monodispersity in a bacterial expression system
Protein Sci., 18, 2009
|
|
4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
1S9A
 
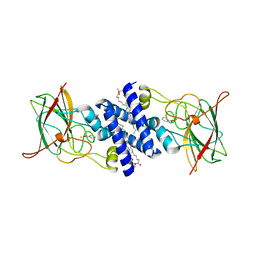 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
1ZG1
 
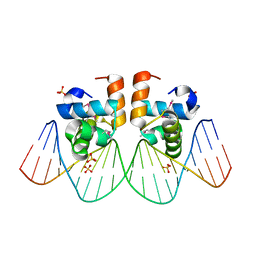 | | NarL complexed to nirB promoter non-palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*GP*GP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*CP*CP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, ... | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
4IBG
 
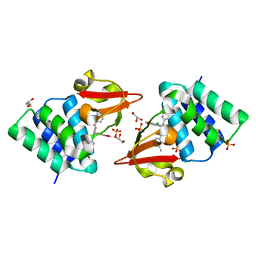 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
1ZGW
 
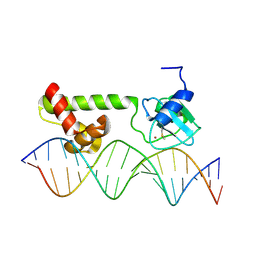 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
4DEA
 
 | | Aurora A in complex with YL1-038-18 | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-(pyrimidine-2,4-diyldiimino)dibenzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3HQT
 
 | | PLP-Dependent Acyl-CoA Transferase CqsA | | Descriptor: | CAI-1 autoinducer synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kelly, R.C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA.
Nat.Chem.Biol., 5, 2009
|
|
3O02
 
 | | The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Chenodeoxycholate | | Descriptor: | CHENODEOXYCHOLIC ACID, Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
6KSP
 
 | |
4QF7
 
 | | Crystal Structure of the Complex of Phospholipase A2 with Corticosterone at 1.48 A Resolution | | Descriptor: | CORTICOSTERONE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
4DHR
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DI5
 
 | |
1ZNJ
 
 | | INSULIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Turkenburg, J.P, Dodson, G.G, Derewenda, U, Smith, G.D, Dodson, E.J, Derewenda, Z.S, Xiao, B. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Refinement of Two Crystal Forms of Native Insulins
To be Published
|
|
5PBC
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09724a | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-imidazole, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4QAB
 
 | | X-RAY STRUCTURE of ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 4-(MORPHOLIN-4-YL)-6-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(morpholin-4-yl)-6-[4-(trifluoromethyl)phenyl]pyrimidin-2-amine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2R6P
 
 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
