5YHZ
 
 | | Structure of Lactococcus lactis ZitR, E41A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ZINC ION, ... | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
6H6P
 
 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
3VHN
 
 | | Y61G mutant of Cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3F07
 
 | | Crystal Structure Analysis of Human HDAC8 complexed with APHA in a new monoclinic crystal form | | Descriptor: | (2E)-N-hydroxy-3-[1-methyl-4-(phenylacetyl)-1H-pyrrol-2-yl]prop-2-enamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
5EGK
 
 | |
6ADX
 
 | | Crystal structure of the Zika virus NS3 helicase (ADP-Mn2+ complex, form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6EOY
 
 | | Transthyretin in complex with 4-(1,3-Benzothiazol-2-yl)-2-methylaniline | | Descriptor: | 4-(1,3-benzothiazol-2-yl)-2-methyl-aniline, Transthyretin | | Authors: | Leite, J.P, Gales, L. | | Deposit date: | 2017-10-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting transthyretin in Alzheimer's disease: Drug discovery of small-molecule chaperones as disease-modifying drug candidates for Alzheimer's disease.
Eur.J.Med.Chem., 226, 2021
|
|
6GIX
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
6AFO
 
 | |
1F0T
 
 | | BOVINE TRYPSIN COMPLEXED WITH RPR131247 | | Descriptor: | 4-HYDROXY-3-[2-OXO-3-(THIENO[3,2-B]PYRIDINE-2-SULFONYLAMINO)-PYRROLIDIN-1-YLMETHYL]-BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Maignan, S, Guilloteau, J.P, Pouzieux, S, Choi-Sledeski, Y.M, Becker, M.R, Klein, S.I, Ewing, W.R, Pauls, H.W, Spada, A.P, Mikol, V. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of human factor Xa complexed with potent inhibitors.
J.Med.Chem., 43, 2000
|
|
5YN8
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6GN0
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNN
 
 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
5YNO
 
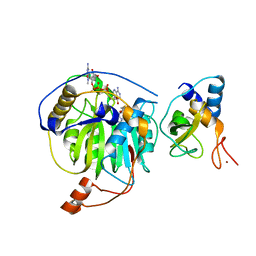 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6GNT
 
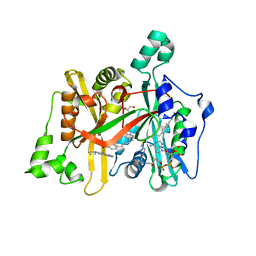 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-(6-pyrrolidin-1-ylquinolin-5-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
1K2F
 
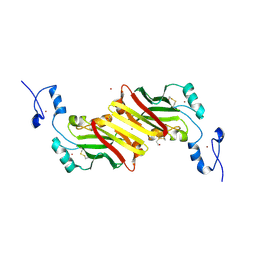 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
5EPM
 
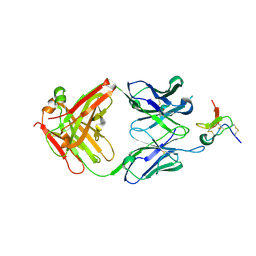 | |
5ENN
 
 | | The crystal structure of Human VPS34 in complex with a selective and potent inhibitor | | Descriptor: | 1-[[4-(cyclopropylmethyl)-5-[2-(pyridin-4-ylamino)pyrimidin-4-yl]pyrimidin-2-yl]amino]-2-methyl-propan-2-ol, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Kearney, E.P. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potent, selective, and orally bioavailable inhibitors of VPS34 provide chemical tools to modulate autophagy in vivo
To Be Published
|
|
6AK7
 
 | | Crystal structure of PPM1K-N94K | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Protein phosphatase 1K, ... | | Authors: | Meisam, R.D, Guo, L.L, Xiao, P. | | Deposit date: | 2018-08-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure of PPM1K-N94K mutant at 2.61 Angstroms resolution
To Be Published
|
|
6ETE
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Crystal structure of KDM4D with tetrazolylhydrazide ligand NR128
To be published
|
|
2TEC
 
 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
5EP1
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | Descriptor: | ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
6EU4
 
 | |
1F4K
 
 | | CRYSTAL STRUCTURE OF THE REPLICATION TERMINATOR PROTEIN/B-SITE DNA COMPLEX | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*AP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*TP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', REPLICATION TERMINATION PROTEIN | | Authors: | Wilce, J.A, Vivian, J.P, Hastings, A.F, Otting, G, Folmer, R.H.A, Duggin, I.G, Wake, R.G, Wilce, M.C.J. | | Deposit date: | 2000-06-08 | | Release date: | 2001-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest
Nat.Struct.Biol., 8, 2001
|
|
1JKH
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
