8VKA
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8ZN2
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
4RSC
 
 | | Crystal structure of RPE65 in complex with emixustat and palmitate | | Descriptor: | (1R)-3-amino-1-[3-(cyclohexylmethoxy)phenyl]propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D, Shi, W, Palczewski, K. | | Deposit date: | 2014-11-07 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic mechanism of a retinoid isomerase essential for vertebrate vision.
Nat.Chem.Biol., 11, 2015
|
|
8VKB
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
9IIL
 
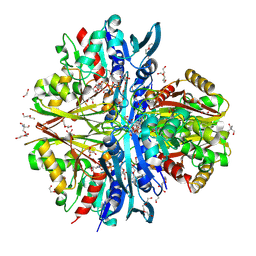 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution
To Be Published
|
|
8XAS
 
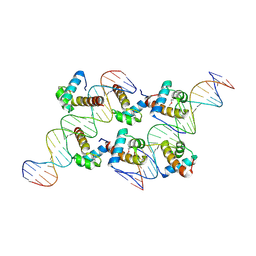 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WIL
 
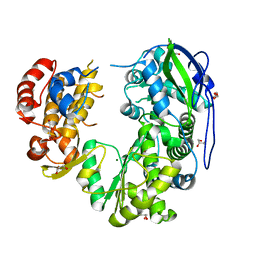 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D55 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
8ZN3
 
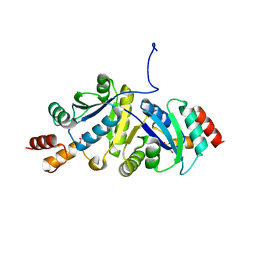 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
4RNE
 
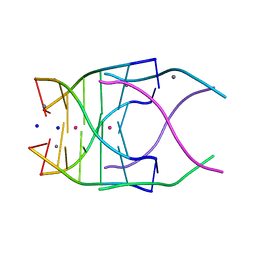 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
4S1G
 
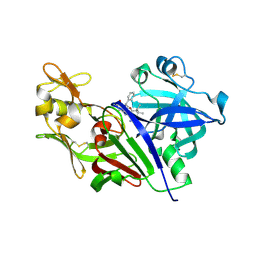 | |
4RVL
 
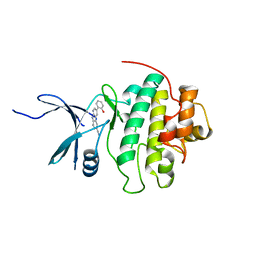 | | CHK1 kinase domain with diazacarbazole compound 7: 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile | | Descriptor: | 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
4RSR
 
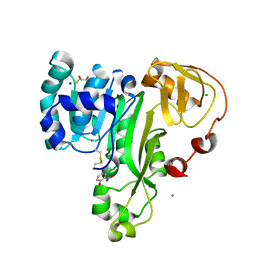 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with trivalent phenyl arsencial derivative-Roxarsone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-arsanyl-2-nitrophenol, Arsenic methyltransferase, ... | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U1I
 
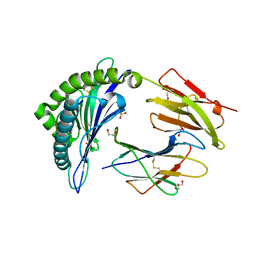 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
8RDK
 
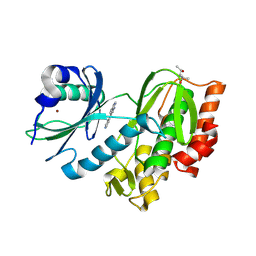 | | Crystal structure of human Haspin (GSG2) kinase bound to MD420 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-pyridin-4-yl-1~{H}-pyrrolo[3,2-g]isoquinoline, Serine/threonine-protein kinase haspin, ... | | Authors: | Kraemer, A, Defois, M, Giraud, F, Moreau, P, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting
Eur.J.Med.Chem., 2024
|
|
4U1C
 
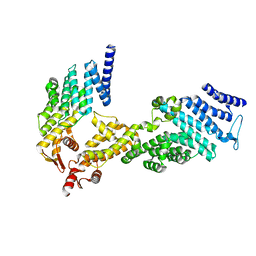 | |
4U1J
 
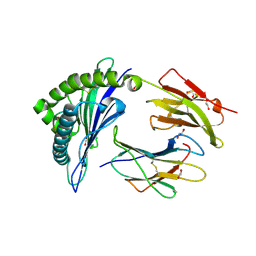 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U36
 
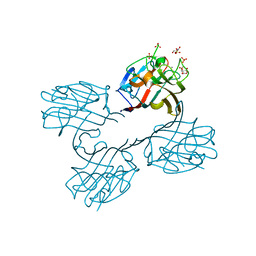 | | Crystal structure of a seed lectin from Vatairea macrocarpa complexed with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4UBO
 
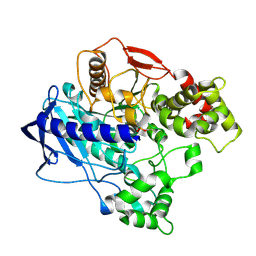 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4U3Z
 
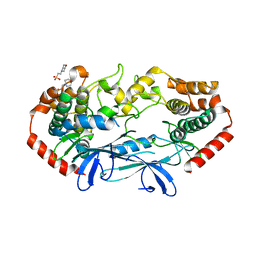 | | APO MAP4K4 T181E Phosphomimetic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 4, SODIUM ION | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U6G
 
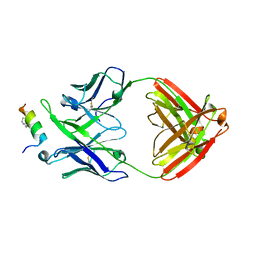 | | Crystal structure of 10E8 Fab in complex with a hydrocarbon-stapled HIV-1 gp41 MPER peptide | | Descriptor: | 10E8 Fab Heavy Chain, 10E8 Fab Light Chain, SAH-MPER(662-683KKK)(B,q) | | Authors: | Ofek, G, Bird, G.H, Walensky, L.D, Kwong, P.D. | | Deposit date: | 2014-07-28 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.2012 Å) | | Cite: | Structural Fidelity and Neutralizing Antibody Binding Capacity of
Hydrocarbon-Stapled HIV-1 Antigens
To be published
|
|
4U40
 
 | |
4U42
 
 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U2A
 
 | | Structure of a lectin from the seeds of Vatairea macrocarpa complexed with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4U0O
 
 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
4UBK
 
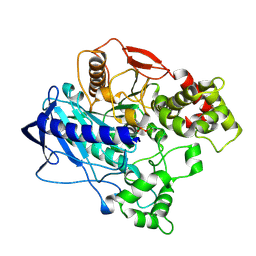 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.40 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
