343D
 
 | |
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
4WVP
 
 | | Crystal structure of an activity-based probe HNE complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BTN-3V3-NLB-OMT-OIC-3V2, ... | | Authors: | Lechtenberg, B.C, Kasperkiewicz, P, Robinson, H.R, Drag, M, Riedl, S.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Elastase-PK101 Structure: Mechanism of an Ultrasensitive Activity-based Probe Revealed.
Acs Chem.Biol., 10, 2015
|
|
1JKU
 
 | | Crystal Structure of Manganese Catalase from Lactobacillus plantarum | | Descriptor: | CALCIUM ION, HYDROXIDE ION, MANGANESE (III) ION, ... | | Authors: | Barynin, V.V, Whittaker, M.M, Antonyuk, S.V, Lamzin, V.S, Harrison, P.M, Artymiuk, P.J, Whittaker, J.W. | | Deposit date: | 2001-07-13 | | Release date: | 2002-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of manganese catalase from Lactobacillus plantarum.
Structure, 9, 2001
|
|
7P4N
 
 | |
1RM4
 
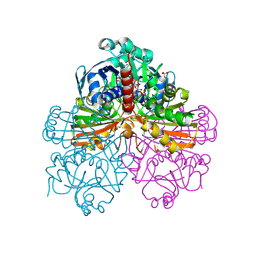 | | Crystal structure of recombinant photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
1PCJ
 
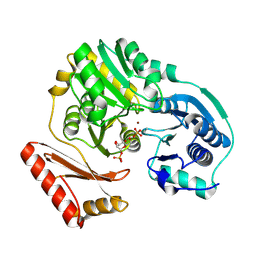 | |
4HHT
 
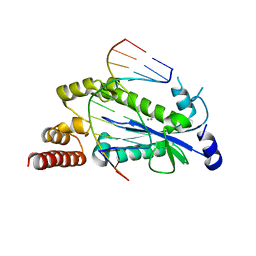 | |
5EXE
 
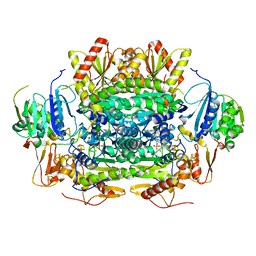 | | Crystal structure of oxalate oxidoreductase from Moorella thermoacetica bound with carboxy-TPP adduct | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | One-carbon chemistry of oxalate oxidoreductase captured by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7PDI
 
 | |
6VCG
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | Descriptor: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5LPN
 
 | | Structure of human Rab10 in complex with the bMERB domain of Mical-1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein-methionine sulfoxide oxidase MICAL1, ... | | Authors: | Rai, A, Oprisko, A, Campos, J, Fu, Y, Friese, T, Itzen, A, Goody, R.S, Mueller, M.P, Gazdag, E.M. | | Deposit date: | 2016-08-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | bMERB domains are bivalent Rab8 family effectors evolved by gene duplication.
Elife, 5, 2016
|
|
1PCM
 
 | |
6ED9
 
 | |
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
6UQC
 
 | | Mouse IgG2a Bispecific Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Tsai, J.C, Davis, J.H, West, S.M, Strop, P. | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and characterization of mouse IgG1 and IgG2a bispecific antibodies for use in syngeneic models.
Mabs, 12, 2019
|
|
1JP3
 
 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
7PN0
 
 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-09-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group
To Be Published
|
|
5LRJ
 
 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin C complex | | Descriptor: | Anabaenopeptin C, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
5LS9
 
 | | Humanized Archaeal ferritin | | Descriptor: | Ferritin, putative, MAGNESIUM ION | | Authors: | Baiocco, P, Trabuco, M.C, Boffi, A. | | Deposit date: | 2016-08-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Humanized archaeal ferritin as a tool for cell targeted delivery.
Nanoscale, 9, 2017
|
|
7PKP
 
 | | NSP2 RNP complex | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DLK
 
 | | Crystal Structure of veratryl alcohol bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dye-decolorizing peroxidase from Bacillus subtilis in complex with veratryl alcohol.
Int.J.Biol.Macromol., 193, 2021
|
|
4Q8S
 
 | | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrophenyl hexadecanoate, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution
To be Published
|
|
4LAJ
 
 | | Crystal structure of HIV-1 YU2 envelope gp120 glycoprotein in complex with CD4-mimetic miniprotein, M48U1, and llama single-domain, broadly neutralizing, co-receptor binding site antibody, JM4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Heavy Chain-Only IgG2b Llama Antibody Effects Near-Pan HIV-1 Neutralization by Recognizing a CD4-Induced Epitope That Includes Elements of Coreceptor- and CD4-Binding Sites.
J.Virol., 87, 2013
|
|
