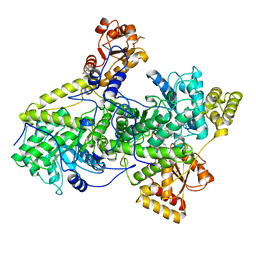
3REQ
 
 | |
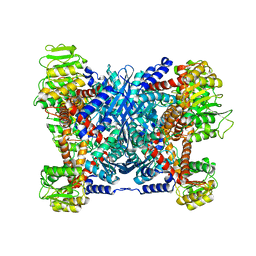
5XVI
 
 | |
7Z4A
 
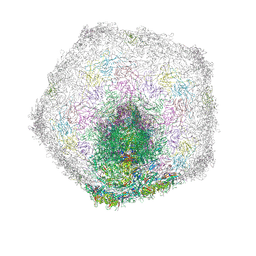 | | Bacteriophage SU10 tail and bottom part of the capsid (C1) | | Descriptor: | Adaptor protein, Major head protein, Portal protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
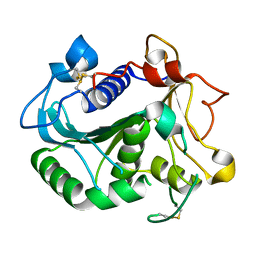
6UNV
 
 | |
6EIX
 
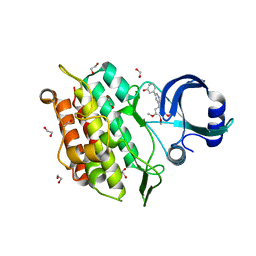 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
6TVF
 
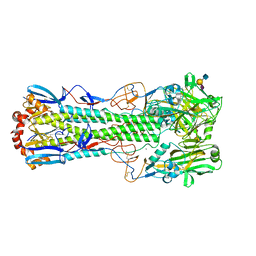 | | Crystal structure of the haemagglutinin from a H10N7 seal influenza virus isolated in Germany in complex with human receptor analogue, 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
7OPX
 
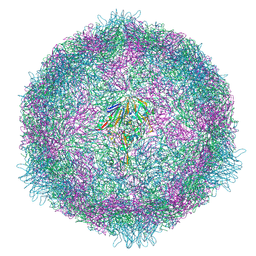 | | CryoEM structure of human enterovirus 70 native virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7Z4W
 
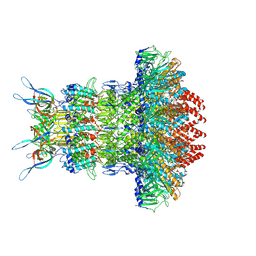 | | gp6/gp15/gp16 connector complex of bacteriophage SPP1 | | Descriptor: | Head completion protein gp15, Head completion protein gp16, Portal protein | | Authors: | Orlov, I, Roche, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2022-03-05 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper.
Nat Commun, 13, 2022
|
|
1BA7
 
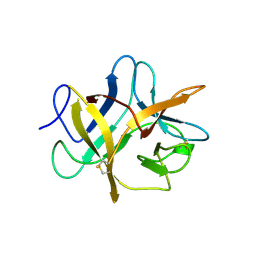 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4QAA
 
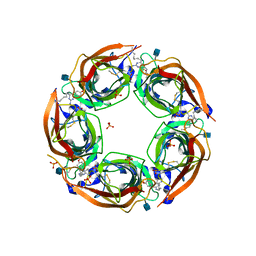 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(4-Methoxyphenyl)-N4-octylpyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~-octylpyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7YXP
 
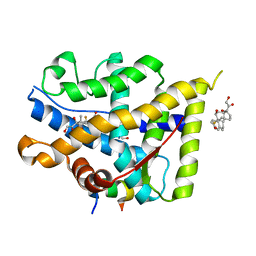 | | Crystal structure of WT AncGR2-LBD WT bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
4BFF
 
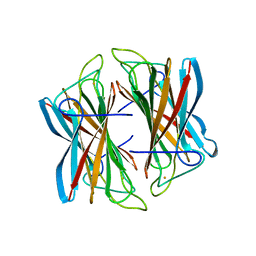 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus in the reduced form | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-03-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
7COW
 
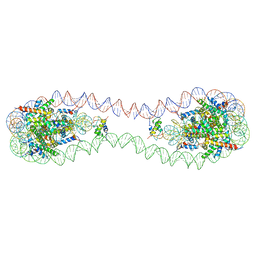 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
5ETU
 
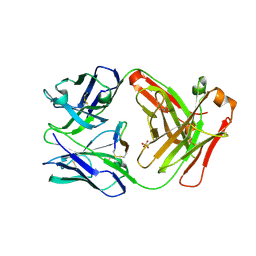 | | Cetuximab Fab in complex with L5E meditope variant | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, L5E meditope variant, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4GLB
 
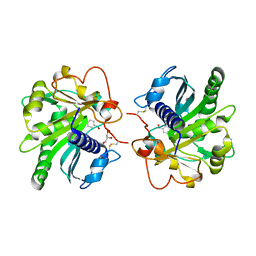 | | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution | | Descriptor: | 4-nitrobenzaldehyde, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution
TO BE PUBLISHED
|
|
5LDO
 
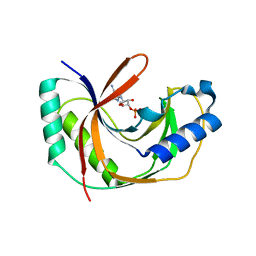 | | Crystal structure of E.coli LigT complexed with 3'-AMP | | Descriptor: | RNA 2',3'-cyclic phosphodiesterase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
4GMJ
 
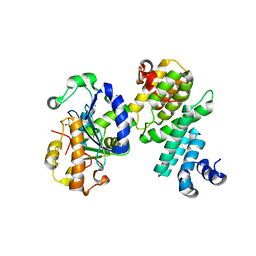 | | Structure of human NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 7, CHLORIDE ION, ... | | Authors: | Petit, P, Weichenrieder, O, Wohlbold, L, Izaurralde, E. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex
Nucleic Acids Res., 40, 2012
|
|
5ZZV
 
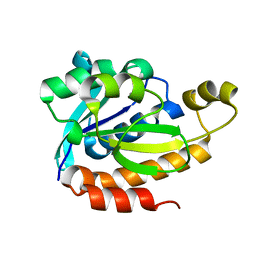 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
4WXG
 
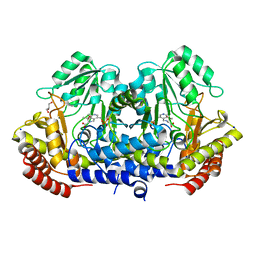 | | Crystal structure of L-Serine Hydroxymethyltransferase in complex with a mixture of L-Threonine and Glycine | | Descriptor: | GLYCEROL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, SODIUM ION, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5LH4
 
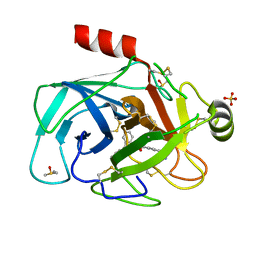 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 1 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-(2,2-diphenylethyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
2MNX
 
 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
5EYR
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 5 at 1.57A resolution | | Descriptor: | 3-[1-[4-(methylaminomethyl)phenyl]piperidin-4-yl]-1-pyrrolidin-1-yl-propan-1-one, EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5LE2
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_15_D12_15_D12 | | Descriptor: | ACETATE ION, DDD_D12_15_D12_15_D12, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEC
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
4WVZ
 
 | | Crystal structure of artificial crosslinked thiol dioxygenase G95C variant from Pseudomonas aeruginosa | | Descriptor: | 3-mercaptopropionate dioxygenase, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-11-09 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Substrate and pH-Dependent Kinetic Profile of 3-Mercaptopropionate Dioxygenase from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
