7LNX
 
 | | I146A mutant of the isopentenyl phosphate kinase from Candidatus methanomethylophilus alvus | | Descriptor: | (2E)-3-methylhept-2-en-1-yl dihydrogen phosphate, (2Z)-3-methylhept-2-en-1-yl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Johnson, B.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
5VJ0
 
 | |
5VJS
 
 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B, a synthetic Zn porphyrin, and Zn(II) cations | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette, ... | | Authors: | Ennist, N.M, Dutton, P.L, Stayrook, S.E, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
7QU9
 
 | | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii. | | Descriptor: | Anthranilate synthase component I family protein, GLYCEROL, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R, Back, C.B, Burton, N.B, Willis, C.L, Stach, J.E.M, Duke, P.W, Hawkins, C. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii.
To Be Published
|
|
5VKB
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and argininamide | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININEAMIDE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7QXV
 
 | | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hemin transport protein | | Authors: | Barker, P.D, Keith, A, Brear, P, Wales, D. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica
To Be Published
|
|
6KCM
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613S with pterin and p-hydrobenzoate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
3HA4
 
 | |
3UM8
 
 | | Wild-type Plasmodium falciparum DHFR-TS complexed with cycloguanil and NADPH | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
5CYO
 
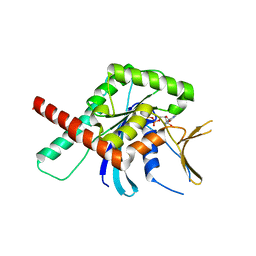 | | High resolution Septin 9 GTPase domain in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-9 | | Authors: | Matos, S.S, Leonardo, D.A, Pereira, H.M, Horjales, E, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2015-07-30 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.0354 Å) | | Cite: | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
6KL8
 
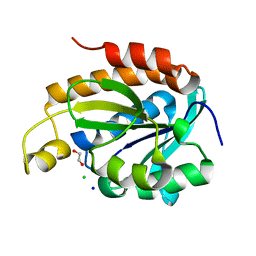 | | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site
To Be Published
|
|
3H7H
 
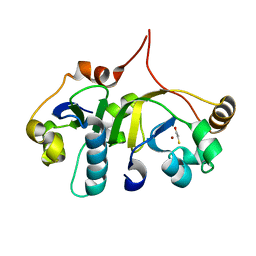 | | Crystal structure of the human transcription elongation factor DSIF, hSpt4/hSpt5 (176-273) | | Descriptor: | BETA-MERCAPTOETHANOL, Transcription elongation factor SPT4, Transcription elongation factor SPT5, ... | | Authors: | Wenzel, S, Wohrl, B.M, Rosch, P, Martins, B.M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface
Biochem.J., 425, 2010
|
|
3HVT
 
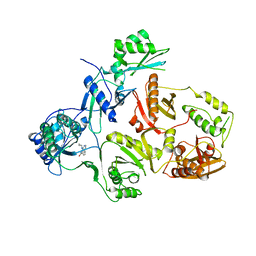 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
5D2H
 
 | |
2GDR
 
 | | Crystal structure of a bacterial glutathione transferase | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Tocheva, E.I, Fortin, P.D, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2006-03-16 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Ternary Complexes of BphK, a Bacterial Glutathione S-Transferase That Reductively Dechlorinates Polychlorinated Biphenyl Metabolites.
J.Biol.Chem., 281, 2006
|
|
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
5VKL
 
 | | SPT6 tSH2-RPB1 1476-1500 pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
4OQP
 
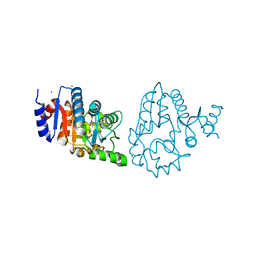 | |
4OIT
 
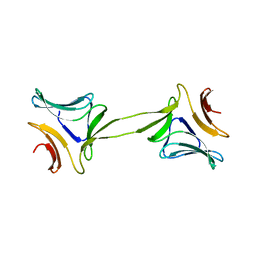 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
4OOA
 
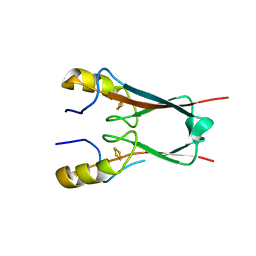 | | CRYSTAL STRUCTURE of NAF1 (MINER1): H114C THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Conlan, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Mittler, R, Jennings, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5W2Y
 
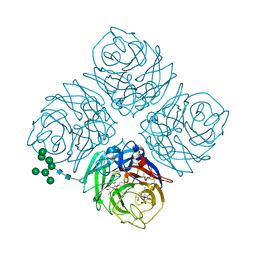 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 9-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-6-[(1R,2R)-1,2-bis(oxidanyl)propyl]-2,3-bis(fluoranyl)-4-oxidanyl-oxane-2-carboxylic acid, (2~{R},3~{R},4~{R},5~{R})-3-acetamido-2-[(1~{R},2~{R})-1,2-bis(oxidanyl)propyl]-5-fluoranyl-4-oxidanyl-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
3V09
 
 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
4O87
 
 | | Crystal structure of a N-tagged Nuclease | | Descriptor: | CITRIC ACID, N-tagged Nuclease, SULFATE ION | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, mechanism, and specificity of a eukaryal tRNA restriction enzyme involved in self-nonself discrimination.
Cell Rep, 7, 2014
|
|
4ODA
 
 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
