8D4Y
 
 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
8BDU
 
 | | H33 variant of DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | PIH1 domain-containing protein 1 | | Authors: | Kolenko, P, Mikulecky, P, Pham, P.N, Schneider, B. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Diffraction anisotropy and paired refinement: crystal structure of H33, a protein binder to interleukin 10.
J.Appl.Crystallogr., 56, 2023
|
|
5IVQ
 
 | |
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
7PGE
 
 | | copper transporter PcoB | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Copper resistance protein B, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, P, Gourdon, P.E. | | Deposit date: | 2021-08-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PcoB is a defense outer membrane protein that facilitates cellular uptake of copper.
Protein Sci., 31, 2022
|
|
6Y2L
 
 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
7NT9
 
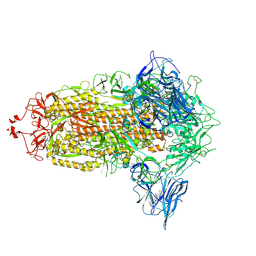 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTA
 
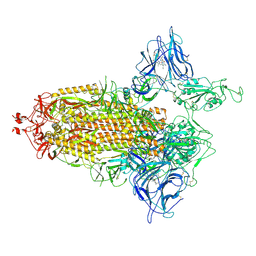 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTC
 
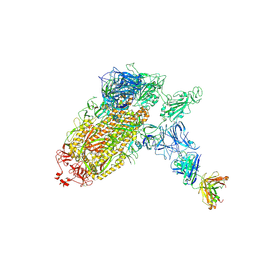 | | Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
8QUB
 
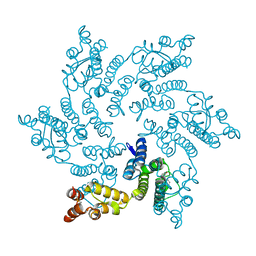 | | Hexameric HIV-1 CA in complex with DDD00074110 | | Descriptor: | (1~{S})-1-phenyl-2,4-dihydro-1~{H}-isoquinolin-3-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUH
 
 | | Hexameric HIV-1 CA in complex with DDD00057456 | | Descriptor: | 4-methylquinolin-2-ol, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
7B4V
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.2, CHLORIDE ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
8QUJ
 
 | | Hexameric HIV-1 CA in complex with DDD00100452 | | Descriptor: | 1,2-ETHANEDIOL, 3-(phenylmethyl)-1~{H}-imidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUI
 
 | | Hexameric HIV-1 CA in complex with DDD00024969 | | Descriptor: | Spacer peptide 1, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUL
 
 | | Hexameric HIV-1 CA in complex with DDD00100555 | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUW
 
 | | Hexameric HIV-1 CA in complex with DDD01044153 | | Descriptor: | (4~{R})-7-oxidanyl-4-phenyl-3,4-dihydro-1~{H}-quinolin-2-one, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUX
 
 | | Hexameric HIV-1 CA in complex with DDD00100333 | | Descriptor: | 1,2-ETHANEDIOL, 4-benzyl-3,4-dihydroquinoxalin-2(1H)-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV1
 
 | | Hexameric HIV-1 CA in complex with DDD01728505 | | Descriptor: | Spacer peptide 1, methyl 2-(2-oxidanylidene-1~{H}-quinolin-4-yl)ethanoate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV9
 
 | | Hexameric HIV-1 CA in complex with DDD01829021 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
3IL8
 
 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6XRL
 
 | | Crystal structure of human PI3K-gamma in complex with inhibitor IPI-549 | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6MQM
 
 | |
8DCJ
 
 | | [A:T] Self-Assembled 3D DNA Rhombohedral Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QUK
 
 | | Hexameric HIV-1 CA in complex with DDD00100439 | | Descriptor: | (phenylmethyl) 3-oxidanylidenepiperazine-1-carboxylate, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
